SEAI 2022 - R - Lab 3
Intro to R
Vincenzo Nardelli - vincnardelli@gmail.com - https://github.com/vincnardelli
Lab structure
Loading and plotting spatial data
library(dplyr)
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
data = read.csv("data/dataNUTS3/dataNUTS3.csv")
head(data)
## NUTS_ID year alpine category pcgdp employ_1564 employ_1564_male
## 1 AT113 1977 No Predominantly rural NA NA NA
## 2 AT112 1977 No Predominantly rural NA NA NA
## 3 AT111 1977 No Predominantly rural NA NA NA
## 4 AT111 1978 No Predominantly rural NA NA NA
## 5 AT112 1978 No Predominantly rural NA NA NA
## 6 AT113 1978 No Predominantly rural NA NA NA
## employ_1564_female employ_5564 unempl educ_2564 fertility pop pop_dens
## 1 NA NA NA NA NA NA NA
## 2 NA NA NA NA NA NA NA
## 3 NA NA NA NA NA NA NA
## 4 NA NA NA NA NA NA NA
## 5 NA NA NA NA NA NA NA
## 6 NA NA NA NA NA NA NA
## ppopGE65 ppopLT15 research_exp state
## 1 NA NA NA AT
## 2 NA NA NA AT
## 3 NA NA NA AT
## 4 NA NA NA AT
## 5 NA NA NA AT
## 6 NA NA NA AT
data <- data %>%
filter(year == 2012)
head(data)
## NUTS_ID year alpine category pcgdp employ_1564 employ_1564_male
## 1 AT111 2012 No Predominantly rural 20600 70.4 75.9
## 2 AT113 2012 No Predominantly rural 22900 70.4 75.9
## 3 AT112 2012 No Predominantly rural 28500 70.4 75.9
## 4 AT127 2012 No Predominantly urban 43200 72.6 77.2
## 5 AT124 2012 No Predominantly rural 25300 72.6 77.2
## 6 AT125 2012 No Predominantly rural 19000 72.6 77.2
## employ_1564_female employ_5564 unempl educ_2564 fertility pop pop_dens
## 1 64.8 38.7 4.6 13.9 1.30 37561 54.1
## 2 64.8 38.7 4.6 13.9 1.30 97721 67.4
## 3 64.8 38.7 4.6 13.9 1.30 150500 99.0
## 4 68.1 42.9 4.6 17.2 1.49 320945 223.1
## 5 68.1 42.9 4.6 17.2 1.49 219354 48.1
## 6 68.1 42.9 4.6 17.2 1.49 123374 51.6
## ppopGE65 ppopLT15 research_exp state
## 1 0.2108836 0.1269934 NA AT
## 2 0.2007347 0.1280994 NA AT
## 3 0.1879601 0.1367442 NA AT
## 4 0.1863808 0.1476172 NA AT
## 5 0.2069577 0.1352015 NA AT
## 6 0.1995396 0.1324752 NA AT
summary(data$category)
## Length Class Mode
## 1474 character character
data <- data %>%
mutate(category = as.factor(category),
state = as.factor(state))
summary(data$category)
## Intermediate Predominantly rural Predominantly urban NA's
## 450 416 261 347
summary(data$state)
## AT BE BG CH CY CZ DE DK EE EL ES FI FR HR HU IE IS IT LI LT
## 35 44 28 26 1 14 396 11 5 52 59 19 101 21 20 8 2 110 1 10
## LU LV ME MK MT NL NO PL PT RO SE SI SK TR UK
## 1 6 1 8 2 40 19 72 25 42 21 12 8 81 173
library(ggplot2)
data <- data %>%
mutate(pop_dens_log = log(pop_dens),
pcgdp_log = log(pcgdp))
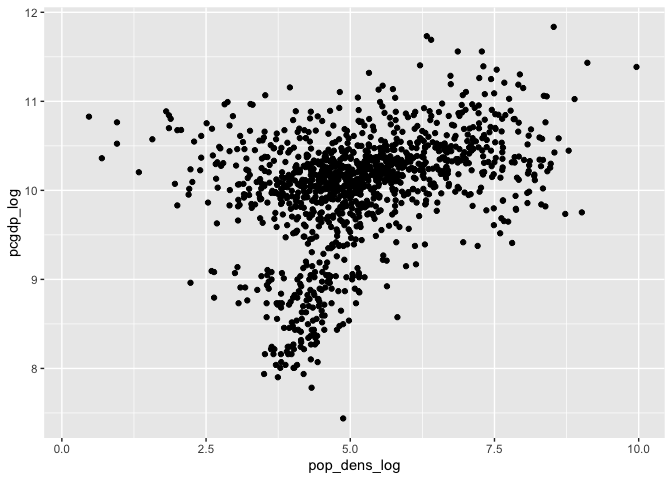
ggplot(data) +
geom_point(aes(pop_dens_log, pcgdp_log))
## Warning: Removed 236 rows containing missing values (geom_point).
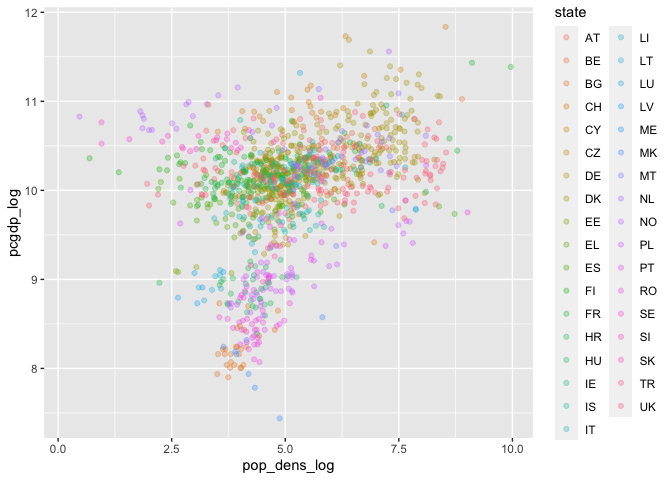
ggplot(data) +
geom_point(aes(pop_dens_log, pcgdp_log, color=state), alpha=0.3)
## Warning: Removed 236 rows containing missing values (geom_point).
Areal data
sf package Link
Package sf represents simple features as records in a data.frame or tibble with a geometry list-column and natively in R all 17 simple feature types for all dimensions (XY, XYZ, XYM, XYZM)
Simple features or simple feature access refers to a formal standard (ISO 19125-1:2004) that describes how objects in the real world can be represented in computers, with emphasis on the spatial geometry of these objects. It also describes how such objects can be stored in and retrieved from databases, and which geometrical operations should be defined for them.
The standard is widely implemented in spatial databases (such as PostGIS), commercial GIS (e.g., ESRI ArcGIS) and forms the vector data basis for libraries such as GDAL. A subset of simple features forms the GeoJSON standard.
R has well-supported classes for storing spatial data (sp) and interfacing to the above mentioned environments (rgdal, rgeos), but has so far lacked a complete implementation of simple features, making conversions at times convoluted, inefficient or incomplete. The package sf tries to fill this gap, and aims at succeeding sp in the long term.
library(sf)
## Linking to GEOS 3.10.2, GDAL 3.4.2, PROJ 8.2.1; sf_use_s2() is TRUE
df_sf <- read_sf("data/dataNUTS3/NUTS_RG_10M_2013.shp")
df_sf
## Simple feature collection with 1951 features and 4 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -63.08826 ymin: -21.38731 xmax: 55.83663 ymax: 71.17354
## Geodetic CRS: ETRS89
## # A tibble: 1,951 × 5
## NUTS_ID STAT_LEVL_ SHAPE_AREA SHAPE_LEN geometry
## <chr> <int> <dbl> <dbl> <MULTIPOLYGON [°]>
## 1 AT 0 10.0 23.9 (((15.15733 48.99178, 15.16025 48.94…
## 2 AT1 1 2.85 11.2 (((15.15733 48.99178, 15.16025 48.94…
## 3 AT11 2 0.473 5.82 (((17.09271 48.09965, 17.06741 48.03…
## 4 AT111 3 0.0866 1.23 (((16.64622 47.4466, 16.57575 47.406…
## 5 AT112 3 0.212 2.81 (((17.1608 48.00666, 17.09466 47.970…
## 6 AT113 3 0.175 2.30 (((16.43376 47.35292, 16.48374 47.28…
## 7 AT12 2 2.33 9.92 (((15.15733 48.99178, 15.16025 48.94…
## 8 AT121 3 0.404 3.75 (((15.50768 48.3196, 15.52095 48.284…
## 9 AT122 3 0.401 3.41 (((16.38889 47.8816, 16.35772 47.866…
## 10 AT123 3 0.148 2.37 (((15.99565 48.14571, 15.91586 48.08…
## # … with 1,941 more rows
df_sf <- df_sf %>%
left_join(data, by="NUTS_ID")
df_sf
## Simple feature collection with 1951 features and 23 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -63.08826 ymin: -21.38731 xmax: 55.83663 ymax: 71.17354
## Geodetic CRS: ETRS89
## # A tibble: 1,951 × 24
## NUTS_ID STAT_LEVL_ SHAPE_AREA SHAPE_LEN geometry year
## <chr> <int> <dbl> <dbl> <MULTIPOLYGON [°]> <int>
## 1 AT 0 10.0 23.9 (((15.15733 48.99178, 15.16025… NA
## 2 AT1 1 2.85 11.2 (((15.15733 48.99178, 15.16025… NA
## 3 AT11 2 0.473 5.82 (((17.09271 48.09965, 17.06741… NA
## 4 AT111 3 0.0866 1.23 (((16.64622 47.4466, 16.57575 … 2012
## 5 AT112 3 0.212 2.81 (((17.1608 48.00666, 17.09466 … 2012
## 6 AT113 3 0.175 2.30 (((16.43376 47.35292, 16.48374… 2012
## 7 AT12 2 2.33 9.92 (((15.15733 48.99178, 15.16025… NA
## 8 AT121 3 0.404 3.75 (((15.50768 48.3196, 15.52095 … 2012
## 9 AT122 3 0.401 3.41 (((16.38889 47.8816, 16.35772 … 2012
## 10 AT123 3 0.148 2.37 (((15.99565 48.14571, 15.91586… 2012
## # … with 1,941 more rows, and 18 more variables: alpine <chr>, category <fct>,
## # pcgdp <dbl>, employ_1564 <dbl>, employ_1564_male <dbl>,
## # employ_1564_female <dbl>, employ_5564 <dbl>, unempl <dbl>, educ_2564 <dbl>,
## # fertility <dbl>, pop <dbl>, pop_dens <dbl>, ppopGE65 <dbl>, ppopLT15 <dbl>,
## # research_exp <dbl>, state <fct>, pop_dens_log <dbl>, pcgdp_log <dbl>
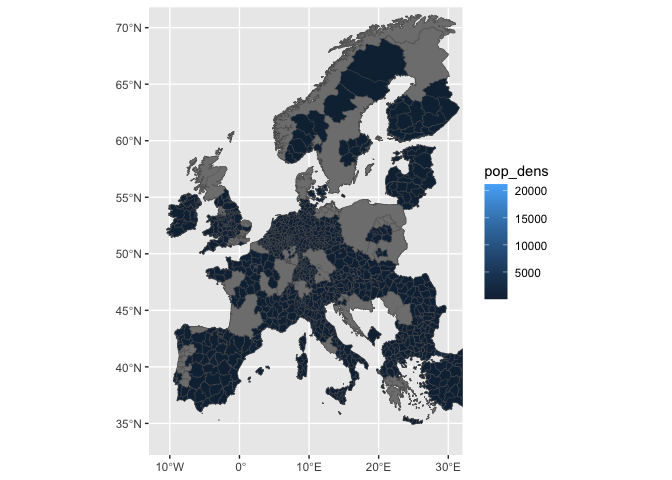
ggplot(df_sf) +
geom_sf(aes(fill=pop_dens))
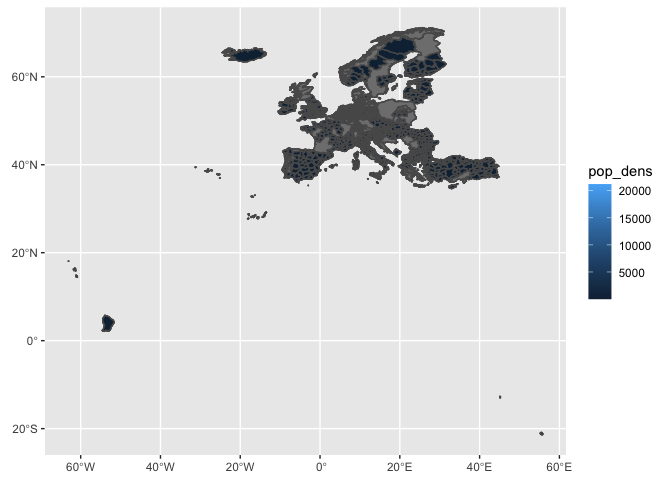
ggplot(df_sf) +
geom_sf(aes(fill=pop_dens), lwd=0.1) +
xlim(-11, 30) +
ylim(34, 70)
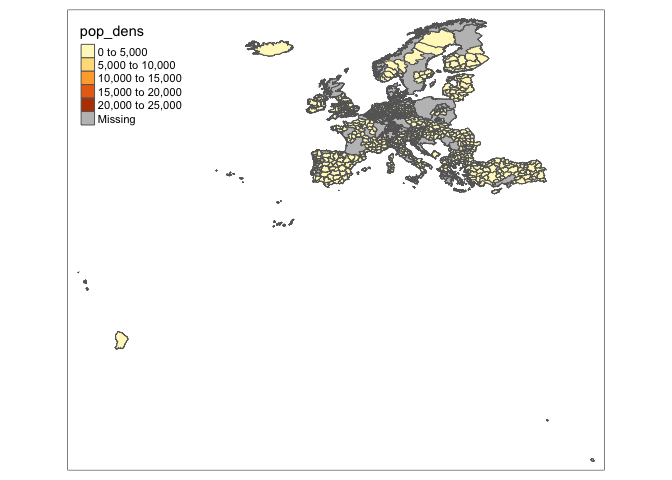
library(tmap)
df_sf <- st_make_valid(df_sf)
tm_shape(df_sf) +
tm_polygons("pop_dens")
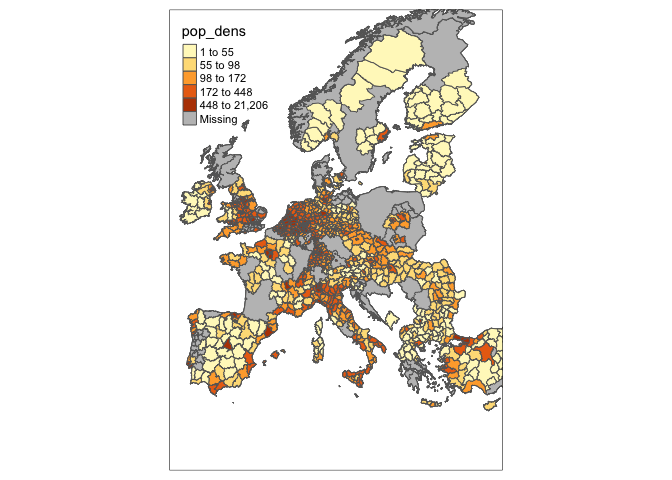
tm_shape(df_sf, bbox=tmaptools::bb(matrix(c(-11,30,34,70),2,2))) +
tm_polygons(col = "pop_dens", style = "quantile")
Interactive mapping
# map <- tm_shape(df_sf, bbox=tmaptools::bb(matrix(c(-11,30,34,70),2,2))) +
# tm_polygons(col = "pop_dens", style = "quantile")
#
# tmap_leaflet(map)
Point data
kc = read_sf("data/kingcounty/kc_house.shp")
kc
## Simple feature collection with 21613 features and 21 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: -122.519 ymin: 47.1559 xmax: -121.315 ymax: 47.7776
## Geodetic CRS: WGS 84
## # A tibble: 21,613 × 22
## id date price bedrooms bathrooms sqft_liv sqft_lot floors waterfront
## <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 7.13e9 2014… 2.22e5 3 1 1180 5650 1 0
## 2 6.41e9 2014… 5.38e5 3 2 2570 7242 2 0
## 3 5.63e9 2015… 1.8 e5 2 1 770 10000 1 0
## 4 2.49e9 2014… 6.04e5 4 3 1960 5000 1 0
## 5 1.95e9 2015… 5.1 e5 3 2 1680 8080 1 0
## 6 7.24e9 2014… 1.23e6 4 4 5420 101930 1 0
## 7 1.32e9 2014… 2.58e5 3 2 1715 6819 2 0
## 8 2.01e9 2015… 2.92e5 3 1 1060 9711 1 0
## 9 2.41e9 2015… 2.30e5 3 1 1780 7470 1 0
## 10 3.79e9 2015… 3.23e5 3 2 1890 6560 2 0
## # … with 21,603 more rows, and 13 more variables: view <dbl>, condition <dbl>,
## # grade <dbl>, sqft_above <dbl>, sqft_basmt <dbl>, yr_built <dbl>,
## # yr_renov <dbl>, zipcode <dbl>, lat <dbl>, long <dbl>, sqft_liv15 <dbl>,
## # sqft_lot15 <dbl>, geometry <POINT [°]>
Coordinate Reference Systems
A coordinate reference system (CRS) then defines how the two-dimensional, projected map in your GIS relates to real places on the earth. The decision of which map projection and CRS to use depends on the regional extent of the area you want to work in, on the analysis you want to do, and often on the availability of data.
https://datacarpentry.org/organization-geospatial/03-crs/
st_crs(kc)
## Coordinate Reference System:
## User input: WGS 84
## wkt:
## GEOGCRS["WGS 84",
## DATUM["World Geodetic System 1984",
## ELLIPSOID["WGS 84",6378137,298.257223563,
## LENGTHUNIT["metre",1]]],
## PRIMEM["Greenwich",0,
## ANGLEUNIT["degree",0.0174532925199433]],
## CS[ellipsoidal,2],
## AXIS["latitude",north,
## ORDER[1],
## ANGLEUNIT["degree",0.0174532925199433]],
## AXIS["longitude",east,
## ORDER[2],
## ANGLEUNIT["degree",0.0174532925199433]],
## ID["EPSG",4326]]
Let’s try to do the same starting from non-projected non-spatial dataset from a csv file!
kc = read.csv("data/kingcounty/kc_house_data.csv")
head(kc)
## id date price bedrooms bathrooms sqft_living sqft_lot
## 1 7129300520 20141013T000000 221900 3 1.00 1180 5650
## 2 6414100192 20141209T000000 538000 3 2.25 2570 7242
## 3 5631500400 20150225T000000 180000 2 1.00 770 10000
## 4 2487200875 20141209T000000 604000 4 3.00 1960 5000
## 5 1954400510 20150218T000000 510000 3 2.00 1680 8080
## 6 7237550310 20140512T000000 1225000 4 4.50 5420 101930
## floors waterfront view condition grade sqft_above sqft_basement yr_built
## 1 1 0 0 3 7 1180 0 1955
## 2 2 0 0 3 7 2170 400 1951
## 3 1 0 0 3 6 770 0 1933
## 4 1 0 0 5 7 1050 910 1965
## 5 1 0 0 3 8 1680 0 1987
## 6 1 0 0 3 11 3890 1530 2001
## yr_renovated zipcode lat long sqft_living15 sqft_lot15
## 1 0 98178 47.5112 -122.257 1340 5650
## 2 1991 98125 47.7210 -122.319 1690 7639
## 3 0 98028 47.7379 -122.233 2720 8062
## 4 0 98136 47.5208 -122.393 1360 5000
## 5 0 98074 47.6168 -122.045 1800 7503
## 6 0 98053 47.6561 -122.005 4760 101930
kc <- kc %>%
st_as_sf(coords = c("long", "lat"), crs = 4326)
ggplot(kc) +
geom_sf(aes(color=price))
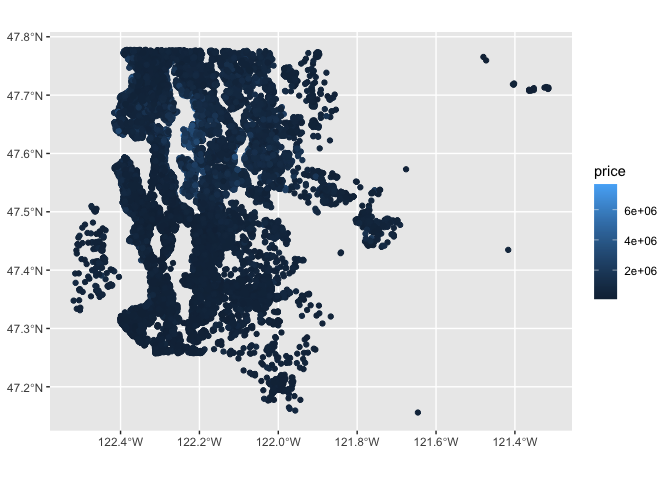
tm_shape(kc) +
tm_bubbles(col="price", scale=0.5, alpha=0.8, style="quantile",
border.lwd=0)
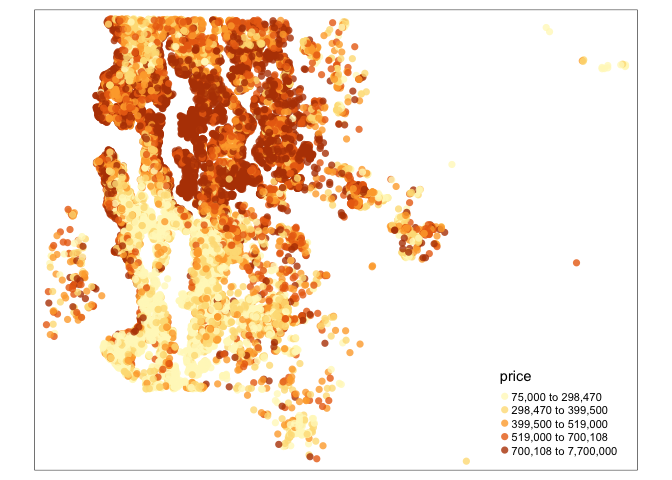
# map <- kc %>%
# head(1000) %>%
# tm_shape() +
# tm_bubbles(col="price", scale=0.2, alpha=0.8, style="quantile",
# border.lwd=0)
#
# tmap_leaflet(map)
Computation of W matrix
spdep package https://r-spatial.github.io/spdep/
W matrix for regular grid data
library(spdep)
## Loading required package: sp
## Loading required package: spData
## To access larger datasets in this package, install the spDataLarge
## package with: `install.packages('spDataLarge',
## repos='https://nowosad.github.io/drat/', type='source')`
n<-4 #the dimension of the grid is n x n
nb_grid <- cell2nb(n, n)
summary(nb_grid)
## Neighbour list object:
## Number of regions: 16
## Number of nonzero links: 48
## Percentage nonzero weights: 18.75
## Average number of links: 3
## Link number distribution:
##
## 2 3 4
## 4 8 4
## 4 least connected regions:
## 1:1 4:1 1:4 4:4 with 2 links
## 4 most connected regions:
## 2:2 3:2 2:3 3:3 with 4 links
xyc <- attr(nb_grid, "region.id")
xy <- matrix(as.integer(unlist(strsplit(xyc, ":"))), ncol=2, byrow=TRUE)
plot(nb_grid, xy)
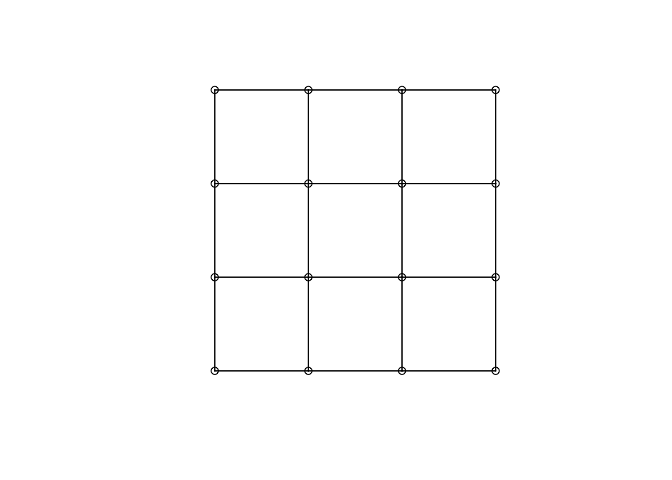
W matrix for irregular data
In the case of irregular data, it is possible to import a GAL (.gal).
#nb <- read.gal("path_to_file")
W matrix from sf
columbus <- read_sf("data/columbus/columbus.shp")
nb<-poly2nb(columbus, queen=T)
nb
## Neighbour list object:
## Number of regions: 49
## Number of nonzero links: 236
## Percentage nonzero weights: 9.829238
## Average number of links: 4.816327
summary(nb)
## Neighbour list object:
## Number of regions: 49
## Number of nonzero links: 236
## Percentage nonzero weights: 9.829238
## Average number of links: 4.816327
## Link number distribution:
##
## 2 3 4 5 6 7 8 9 10
## 5 9 12 5 9 3 4 1 1
## 5 least connected regions:
## 1 6 42 46 47 with 2 links
## 1 most connected region:
## 20 with 10 links
nb[[1]]
## [1] 2 3
columbus_sp <- as(columbus, 'Spatial')
nb_sf <- as(nb2lines(nb, coords = coordinates(columbus_sp)), 'sf')
## Warning in CRS(proj4string): CRS: projargs should not be NULL; set to NA
nb_sf <- st_set_crs(nb_sf, st_crs(columbus))
ggplot(columbus) +
geom_sf(fill = 'gray', color = 'white') +
geom_sf(data = nb_sf) +
theme_minimal() +
ylab("Latitude") +
xlab("Longitude")
 List
W object it resume a W matrix, saving space. A listw object has 3
components: 1) a nb object 2)list of n numeric vectors, each of the same
length as the corresponding non-zero vectors in the nb object. These
give the values of the spatial weights for each i-j neighbour pair.
3)the style of W as a character code: “B” for binary weights taking
values zero or one, “W” for row-standardized matrix,“C” is globally
standardised
List
W object it resume a W matrix, saving space. A listw object has 3
components: 1) a nb object 2)list of n numeric vectors, each of the same
length as the corresponding non-zero vectors in the nb object. These
give the values of the spatial weights for each i-j neighbour pair.
3)the style of W as a character code: “B” for binary weights taking
values zero or one, “W” for row-standardized matrix,“C” is globally
standardised
listw <- nb2listw(nb, style="W")
listw
## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 49
## Number of nonzero links: 236
## Percentage nonzero weights: 9.829238
## Average number of links: 4.816327
##
## Weights style: W
## Weights constants summary:
## n nn S0 S1 S2
## W 49 2401 49 22.75119 203.7091
nb2listw(nb, style="B")
## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 49
## Number of nonzero links: 236
## Percentage nonzero weights: 9.829238
## Average number of links: 4.816327
##
## Weights style: B
## Weights constants summary:
## n nn S0 S1 S2
## B 49 2401 236 472 5304
W matrix for point data
boston <- read_sf("data/boston/boston.shp")
coord <- cbind(boston$LON,boston$LAT)
head(coord)
## [,1] [,2]
## [1,] -70.9550 42.2550
## [2,] -70.9500 42.2875
## [3,] -70.9360 42.2830
## [4,] -70.9280 42.2930
## [5,] -70.9220 42.2980
## [6,] -70.9165 42.3040
KNN
knn = knearneigh(coord, k=5)
nb = knn2nb(knn)
plot(nb, coord)
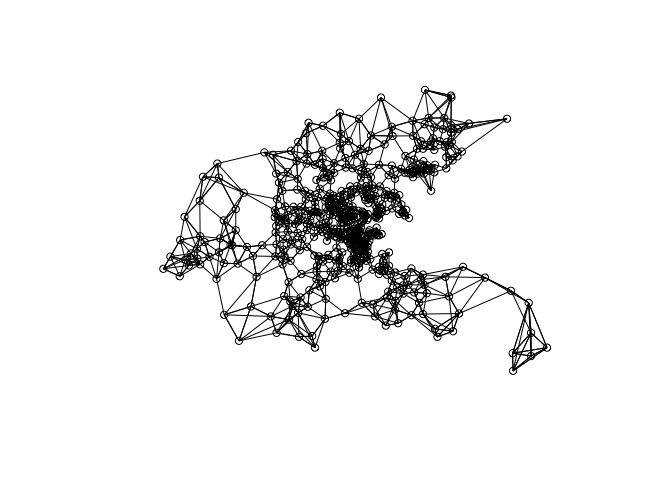
listw <- nb2listw(nb)
listw
## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 506
## Number of nonzero links: 2530
## Percentage nonzero weights: 0.9881423
## Average number of links: 5
## Non-symmetric neighbours list
##
## Weights style: W
## Weights constants summary:
## n nn S0 S1 S2
## W 506 256036 506 178 2087.92
Distance
distM<- as.matrix(dist(coord))
distM[1:5, 1:5]
## 1 2 3 4 5
## 1 0.00000000 0.03288237 0.03383785 0.04661545 0.05420332
## 2 0.03288237 0.00000000 0.01470544 0.02267708 0.02990401
## 3 0.03383785 0.01470544 0.00000000 0.01280625 0.02051828
## 4 0.04661545 0.02267708 0.01280625 0.00000000 0.00781025
## 5 0.05420332 0.02990401 0.02051828 0.00781025 0.00000000
W1<- 1/(distM)
diag(W1) <- 0
W2<- 1/(1+distM)^2
diag(W2) <- 0
W3<- exp(-distM^2)
diag(W3) <- 0
mat2listw(W1)
## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 506
## Number of nonzero links: 255530
## Percentage nonzero weights: 99.80237
## Average number of links: 505
##
## Weights style: M
## Weights constants summary:
## n nn S0 S1 S2
## M 506 256036 4006196 414055665 149609290044
Threshold distance
nb <- dnearneigh(coord, 0, 1.5, longlat = T)
nb
## Neighbour list object:
## Number of regions: 506
## Number of nonzero links: 6648
## Percentage nonzero weights: 2.59651
## Average number of links: 13.13834
## 49 regions with no links:
## 1 41 55 56 65 66 67 68 196 197 198 199 200 201 203 204 205 253 254 255
## 256 257 274 284 285 286 287 288 290 292 300 302 303 304 331 336 342 343
## 344 345 348 349 350 351 352 353 354 355 356
plot(nb, coord)
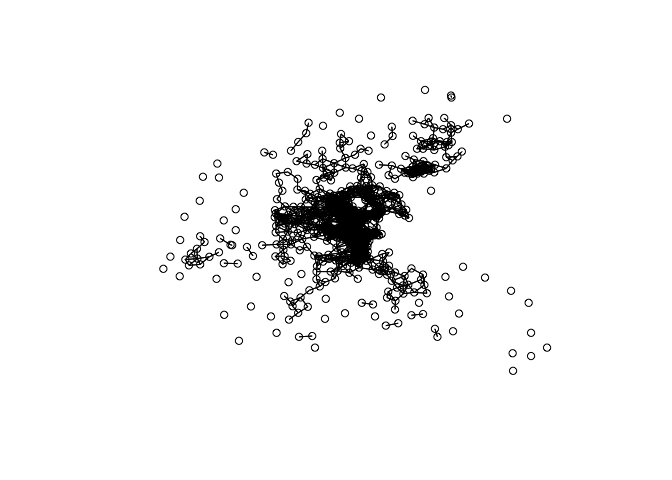
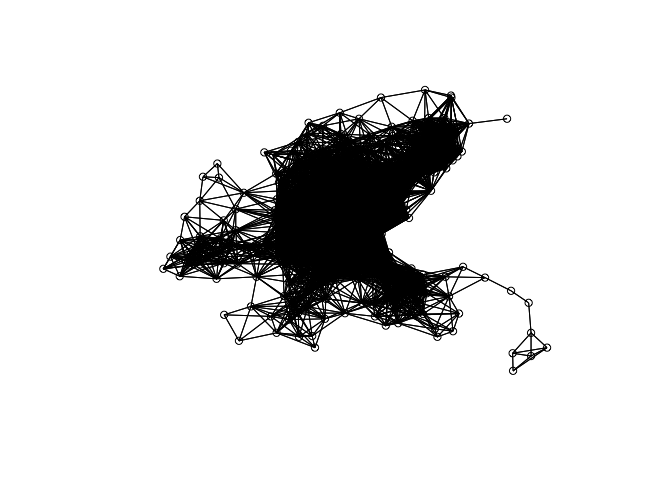
nb <- dnearneigh(coord, 0, 5, longlat = T)
nb
## Neighbour list object:
## Number of regions: 506
## Number of nonzero links: 51400
## Percentage nonzero weights: 20.0753
## Average number of links: 101.581
listw <- nb2listw(nb)
listw
## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 506
## Number of nonzero links: 51400
## Percentage nonzero weights: 20.0753
## Average number of links: 101.581
##
## Weights style: W
## Weights constants summary:
## n nn S0 S1 S2
## W 506 256036 506 29.0168 2042.721
plot(nb, coord)