SEAI 2022 - R - Lab 5
Spatial Autocorrelation with R
Vincenzo Nardelli - vincnardelli@gmail.com - https://github.com/vincnardelli
Lab structure
Let’s load columbus data
library(sf)
## Linking to GEOS 3.10.2, GDAL 3.4.2, PROJ 8.2.1; sf_use_s2() is TRUE
library(dplyr)
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
library(tmap)
library(spdep)
## Loading required package: sp
## Loading required package: spData
## To access larger datasets in this package, install the spDataLarge
## package with: `install.packages('spDataLarge',
## repos='https://nowosad.github.io/drat/', type='source')`
library(ggplot2)
columbus <- read_sf("data/columbus/columbus.shp")
tm_shape(columbus) +
tm_polygons("CRIME")
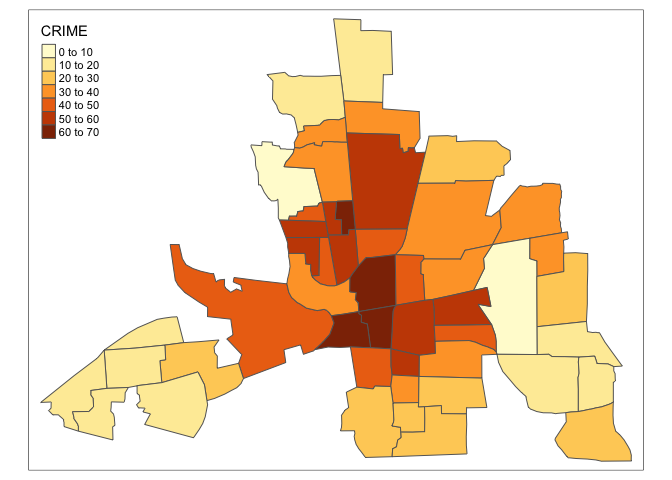
tm_shape(columbus) +
tm_polygons("CRIME")
nb<-poly2nb(columbus, queen=T)
nb
## Neighbour list object:
## Number of regions: 49
## Number of nonzero links: 236
## Percentage nonzero weights: 9.829238
## Average number of links: 4.816327
listw <- nb2listw(nb)
listw
## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 49
## Number of nonzero links: 236
## Percentage nonzero weights: 9.829238
## Average number of links: 4.816327
##
## Weights style: W
## Weights constants summary:
## n nn S0 S1 S2
## W 49 2401 49 22.75119 203.7091
columbus_sp <- as(columbus, 'Spatial')
nb_sf <- as(nb2lines(nb, coords = coordinates(columbus_sp)), 'sf')
## Warning in CRS(proj4string): CRS: projargs should not be NULL; set to NA
nb_sf <- st_set_crs(nb_sf, st_crs(columbus))
ggplot(columbus) +
geom_sf(fill = 'gray', color = 'white') +
geom_sf(data = nb_sf) +
theme_minimal() +
ylab("Latitude") +
xlab("Longitude")

Spatial lag
Compute the lagged value from the W matrix
yla**gi = ∑jwi, jyj
columbus$CRIME
## [1] 15.725980 18.801754 30.626781 32.387760 50.731510 26.066658 0.178269
## [8] 38.425858 30.515917 34.000835 62.275448 56.705669 46.716129 57.066132
## [15] 48.585487 54.838711 36.868774 43.962486 54.521965 0.223797 40.074074
## [22] 33.705048 20.048504 38.297871 61.299175 40.969742 52.794430 56.919785
## [29] 60.750446 68.892044 17.677214 19.145592 41.968163 23.974028 39.175053
## [36] 14.305556 42.445076 53.710938 19.100863 16.241299 18.905146 16.491890
## [43] 36.663612 25.962263 29.028488 16.530533 27.822861 26.645266 22.541491
columbus$CRIME_lag <- lag.listw(listw, columbus$CRIME)
columbus$CRIME_lag
## [1] 24.71427 26.24684 29.41175 34.64648 40.46533 40.62371 49.72845 41.49913
## [9] 36.94778 25.32838 49.85745 43.25009 38.09398 42.82054 50.10508 52.36547
## [17] 18.09105 51.18117 46.44216 32.44636 43.72131 35.90408 18.74605 54.90556
## [25] 49.97125 48.62931 33.20420 47.42423 51.79066 48.57333 19.12682 13.85469
## [33] 37.27827 22.13718 33.09855 18.75491 50.99422 43.65271 16.17110 21.05449
## [41] 21.06992 19.13979 36.16118 35.74727 32.07386 16.70321 17.57322 28.54896
## [49] 27.21201
listw2mat(listw) %*% columbus$CRIME
## [,1]
## 1 24.71427
## 2 26.24684
## 3 29.41175
## 4 34.64648
## 5 40.46533
## 6 40.62371
## 7 49.72845
## 8 41.49913
## 9 36.94778
## 10 25.32838
## 11 49.85745
## 12 43.25009
## 13 38.09398
## 14 42.82054
## 15 50.10508
## 16 52.36547
## 17 18.09105
## 18 51.18117
## 19 46.44216
## 20 32.44636
## 21 43.72131
## 22 35.90408
## 23 18.74605
## 24 54.90556
## 25 49.97125
## 26 48.62931
## 27 33.20420
## 28 47.42423
## 29 51.79066
## 30 48.57333
## 31 19.12682
## 32 13.85469
## 33 37.27827
## 34 22.13718
## 35 33.09855
## 36 18.75491
## 37 50.99422
## 38 43.65271
## 39 16.17110
## 40 21.05449
## 41 21.06992
## 42 19.13979
## 43 36.16118
## 44 35.74727
## 45 32.07386
## 46 16.70321
## 47 17.57322
## 48 28.54896
## 49 27.21201
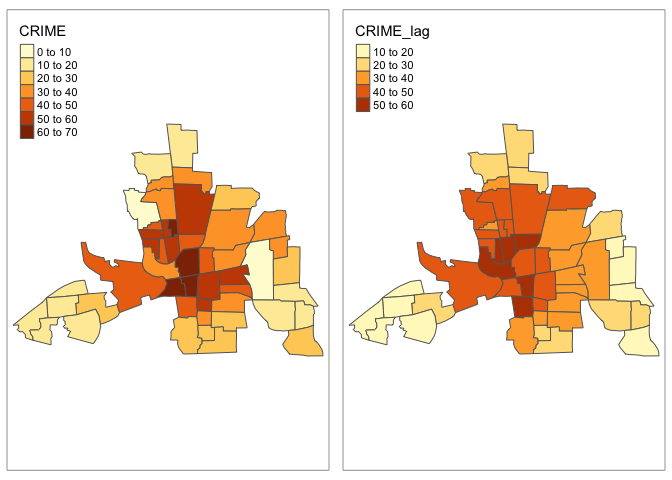
m1 <- tm_shape(columbus) +
tm_polygons("CRIME")
m2 <- tm_shape(columbus) +
tm_polygons("CRIME_lag")
tmap_arrange(m1, m2)
Moran’s Index
moran.test(columbus$CRIME, listw)
##
## Moran I test under randomisation
##
## data: columbus$CRIME
## weights: listw
##
## Moran I statistic standard deviate = 5.5894, p-value = 1.139e-08
## alternative hypothesis: greater
## sample estimates:
## Moran I statistic Expectation Variance
## 0.500188557 -0.020833333 0.008689289
moran.mc(columbus$CRIME, listw, nsim=1000)
##
## Monte-Carlo simulation of Moran I
##
## data: columbus$CRIME
## weights: listw
## number of simulations + 1: 1001
##
## statistic = 0.50019, observed rank = 1001, p-value = 0.000999
## alternative hypothesis: greater
moran.plot(columbus$CRIME, listw)
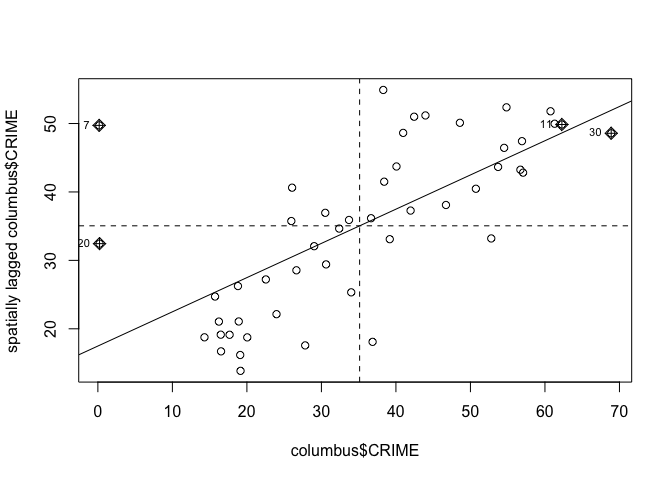
mp <- moran.plot(columbus$CRIME, listw)
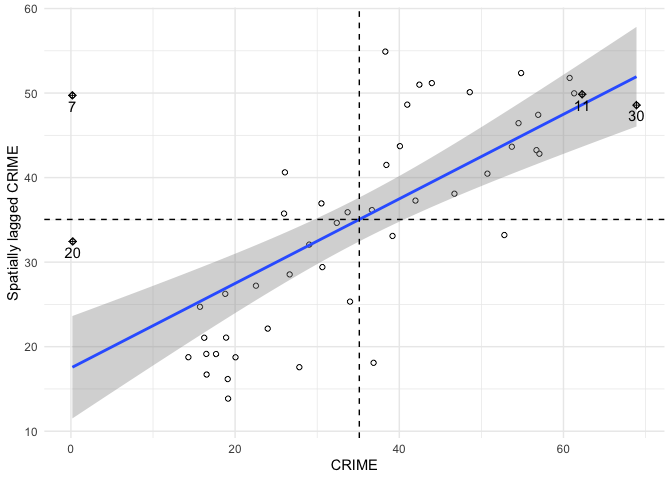
xname <- "CRIME"
ggplot(mp, aes(x=x, y=wx)) + geom_point(shape=1) +
geom_smooth(formula=y ~ x, method="lm") +
geom_hline(yintercept=mean(mp$wx), lty=2) +
geom_vline(xintercept=mean(mp$x), lty=2) + theme_minimal() +
geom_point(data=mp[mp$is_inf,], aes(x=x, y=wx), shape=9) +
geom_text(data=mp[mp$is_inf,], aes(x=x, y=wx, label=labels, vjust=1.5)) +
xlab(xname) + ylab(paste0("Spatially lagged ", xname))
 ##
Local Moran
##
Local Moran
locm <- localmoran_perm(columbus$CRIME, listw)
columbus <- columbus %>%
mutate(CRIME_lag = lag.listw(listw, columbus$CRIME),
p_value = locm[, 5],
cluster = case_when(p_value < 0.05 & CRIME > mean(CRIME) & CRIME_lag > mean(CRIME_lag) ~ "HH",
p_value < 0.05 & CRIME < mean(CRIME) & CRIME_lag < mean(CRIME_lag) ~ "LL",
p_value < 0.05 & CRIME > mean(CRIME) & CRIME_lag < mean(CRIME_lag) ~ "HL",
p_value < 0.05 & CRIME < mean(CRIME) & CRIME_lag > mean(CRIME_lag) ~ "LH"),
cluster = factor(cluster, levels = c("HH", "LL", "HL", "LH")))
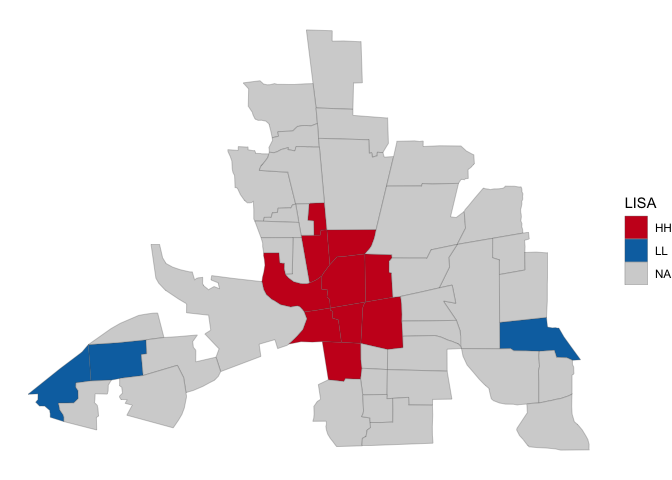
lisa_palette <- c("#ca0020","#0571b0","#f4a582","#92c5de")
ggplot(columbus) +
geom_sf(aes(fill=cluster), lwd=0.1) +
theme_void() +
scale_fill_manual(na.value = "lightgray", name="LISA",
values = (lisa_palette))