SEAI 2022 - Python - Lab 7
ESDA & Spatial Econometrics Models
Vincenzo Nardelli - vincnardelli@gmail.com - https://github.com/vincnardelli
Before to start… install Python Libraries!
! pip install pysal
! pip install cartoframes
! pip install --user urllib3==1.22
Collecting pysal
Downloading https://files.pythonhosted.org/packages/c0/3e/179620647a54eef9ef8173ee82a42b0e33b1e80bbce21998ba8ffc62a0f7/pysal-2.4.0.tar.gz
Collecting libpysal>=4.4.0
[?25l Downloading https://files.pythonhosted.org/packages/cc/e9/36be6aeea8b6d6ac797c656e1f80bdaddc075d049433495680432d9aedd9/libpysal-4.4.0-py3-none-any.whl (2.4MB)
[K |████████████████████████████████| 2.4MB 33.2MB/s
[?25hCollecting access>=1.1.3
Downloading https://files.pythonhosted.org/packages/41/50/d7fb5466976af95e4c3481f8408f29744e9411e46a3f9d610a95e741f57b/access-1.1.3-py3-none-any.whl
Collecting esda>=2.3.6
[?25l Downloading https://files.pythonhosted.org/packages/20/02/7c36f3c7c22f048f50ada6402ff3fbe51e0c5365f7d0227196b84d43c82b/esda-2.3.6-py3-none-any.whl (102kB)
[K |████████████████████████████████| 112kB 58.0MB/s
[?25hCollecting giddy>=2.3.3
[?25l Downloading https://files.pythonhosted.org/packages/5c/71/f7fc2b41e36b2a06bac30937435d2dedd125f74f3157213ec121ff52d961/giddy-2.3.3-py3-none-any.whl (60kB)
[K |████████████████████████████████| 61kB 8.6MB/s
[?25hCollecting inequality>=1.0.0
Downloading https://files.pythonhosted.org/packages/74/0f/9ed2d097f29160d0c873f33ffc0b9806c1083e3611acb2143eb66adcf580/inequality-1.0.0.tar.gz
Collecting pointpats>=2.2.0
[?25l Downloading https://files.pythonhosted.org/packages/5f/2b/a3e99c42b37c3e848eb8e55f2ee2b2d281ae0d49df261f13eaaeba7171a5/pointpats-2.2.0.tar.gz (55kB)
[K |████████████████████████████████| 61kB 9.4MB/s
[?25hCollecting segregation>=1.5.0
[?25l Downloading https://files.pythonhosted.org/packages/21/82/db8d8f2c25d52e774dcdc65861853f87566c7512204764d1b34acf2d7e50/segregation-1.5.0-py3-none-any.whl (82kB)
[K |████████████████████████████████| 92kB 11.8MB/s
[?25hCollecting spaghetti>=1.5.6
[?25l Downloading https://files.pythonhosted.org/packages/cc/fc/3404a77c81b217f5daece7cea5efd1624852b71f225964e2e8bec6bb4ac2/spaghetti-1.6.0-py3-none-any.whl (45kB)
[K |████████████████████████████████| 51kB 7.4MB/s
[?25hCollecting mgwr>=2.1.2
[?25l Downloading https://files.pythonhosted.org/packages/8e/14/e18342b62655ec7f37624ff936d6f273f82b51913cd558110fd4d670794e/mgwr-2.1.2.tar.gz (41kB)
[K |████████████████████████████████| 51kB 8.4MB/s
[?25hCollecting spglm>=1.0.8
Downloading https://files.pythonhosted.org/packages/07/8f/03f07807967595d2632dbe0f7c442511a5ff422103b49c76557f9eed40c5/spglm-1.0.8.tar.gz
Collecting spint>=1.0.7
Downloading https://files.pythonhosted.org/packages/7e/c5/e4862ab3f745a1886135b07e498e77504c14e468d0a6b89f7270f5def979/spint-1.0.7.tar.gz
Collecting spreg>=1.2.2
[?25l Downloading https://files.pythonhosted.org/packages/40/2d/b6559677b66b2f228fdf02fdb5e7da3466057f9e2c696bea7704138ae346/spreg-1.2.2-py3-none-any.whl (209kB)
[K |████████████████████████████████| 215kB 52.3MB/s
[?25hCollecting spvcm>=0.3.0
[?25l Downloading https://files.pythonhosted.org/packages/b4/c9/43fb98bc60728b76fceee175119a1ff7e5033ed9012d07570e34a2a19a8f/spvcm-0.3.0.tar.gz (5.7MB)
[K |████████████████████████████████| 5.7MB 38.5MB/s
[?25hCollecting tobler>=0.6.0
Downloading https://files.pythonhosted.org/packages/5c/88/93acf0ad7c724565975924649928ea9db00e8d94279eb40c7819708b6105/tobler-0.7.0-py3-none-any.whl
Collecting mapclassify>=2.4.2
Downloading https://files.pythonhosted.org/packages/22/8e/d968c0945d41bb02de0efaa92e31e43a817dc52d30e82b4dfdda407a1903/mapclassify-2.4.2-py3-none-any.whl
Collecting splot>=1.1.3
Downloading https://files.pythonhosted.org/packages/83/1c/afb3e3eeeda4eef065f0b3249ca74cbcc44af3bc59ddb5a21b5445753955/splot-1.1.3.tar.gz
Collecting spopt>=0.1.1
[?25l Downloading https://files.pythonhosted.org/packages/a9/df/c8ce65a07f7ab1e69b721533b950dd9854f2f7e6a472eca60c13f5770e24/spopt-0.1.1-py3-none-any.whl (54kB)
[K |████████████████████████████████| 61kB 8.7MB/s
[?25hCollecting urllib3>=1.26
[?25l Downloading https://files.pythonhosted.org/packages/0c/cd/1e2ec680ec7b09846dc6e605f5a7709dfb9d7128e51a026e7154e18a234e/urllib3-1.26.5-py2.py3-none-any.whl (138kB)
[K |████████████████████████████████| 143kB 62.8MB/s
[?25hCollecting python-dateutil<=2.8.0
[?25l Downloading https://files.pythonhosted.org/packages/41/17/c62faccbfbd163c7f57f3844689e3a78bae1f403648a6afb1d0866d87fbb/python_dateutil-2.8.0-py2.py3-none-any.whl (226kB)
[K |████████████████████████████████| 235kB 61.9MB/s
[?25hRequirement already satisfied: pytest in /usr/local/lib/python3.7/dist-packages (from pysal) (3.6.4)
Collecting pytest-cov
Downloading https://files.pythonhosted.org/packages/ba/84/576b071aef9ac9301e5c0ff35d117e12db50b87da6f12e745e9c5f745cc2/pytest_cov-2.12.1-py2.py3-none-any.whl
Requirement already satisfied: coverage in /usr/local/lib/python3.7/dist-packages (from pysal) (3.7.1)
Requirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from libpysal>=4.4.0->pysal) (2.23.0)
Requirement already satisfied: beautifulsoup4 in /usr/local/lib/python3.7/dist-packages (from libpysal>=4.4.0->pysal) (4.6.3)
Requirement already satisfied: jinja2 in /usr/local/lib/python3.7/dist-packages (from libpysal>=4.4.0->pysal) (2.11.3)
Requirement already satisfied: scipy>=0.11 in /usr/local/lib/python3.7/dist-packages (from libpysal>=4.4.0->pysal) (1.4.1)
Requirement already satisfied: numpy>=1.3 in /usr/local/lib/python3.7/dist-packages (from libpysal>=4.4.0->pysal) (1.19.5)
Requirement already satisfied: pandas in /usr/local/lib/python3.7/dist-packages (from libpysal>=4.4.0->pysal) (1.1.5)
Requirement already satisfied: scikit-learn in /usr/local/lib/python3.7/dist-packages (from esda>=2.3.6->pysal) (0.22.2.post1)
Collecting quantecon>=0.4.7
[?25l Downloading https://files.pythonhosted.org/packages/7e/6b/e78ec352947208c6172e50dcb086f95f7b63eb5204f26f46b676ed5bb31c/quantecon-0.5.0-py3-none-any.whl (229kB)
[K |████████████████████████████████| 235kB 44.8MB/s
[?25hRequirement already satisfied: matplotlib in /usr/local/lib/python3.7/dist-packages (from pointpats>=2.2.0->pysal) (3.2.2)
Collecting opencv-contrib-python>=4.2.0
[?25l Downloading https://files.pythonhosted.org/packages/27/f5/21227eb87cd5990a0f4235d8264db7d8f62f6a9ccbf2cd2c367f7d903cd2/opencv_contrib_python-4.5.2.54-cp37-cp37m-manylinux2014_x86_64.whl (57.4MB)
[K |████████████████████████████████| 57.4MB 74kB/s
[?25hRequirement already satisfied: tqdm in /usr/local/lib/python3.7/dist-packages (from segregation>=1.5.0->pysal) (4.41.1)
Requirement already satisfied: seaborn in /usr/local/lib/python3.7/dist-packages (from segregation>=1.5.0->pysal) (0.11.1)
Collecting geopandas
[?25l Downloading https://files.pythonhosted.org/packages/d7/bf/e9cefb69d39155d122b6ddca53893b61535fa6ffdad70bf5ef708977f53f/geopandas-0.9.0-py2.py3-none-any.whl (994kB)
[K |████████████████████████████████| 1.0MB 31.4MB/s
[?25hCollecting rtree
[?25l Downloading https://files.pythonhosted.org/packages/51/05/5a67111cee91d2165a2bcb855f442186e3d76ddef834596cc84d4875c401/Rtree-0.9.7-cp37-cp37m-manylinux2010_x86_64.whl (994kB)
[K |████████████████████████████████| 1.0MB 34.3MB/s
[?25hCollecting pygeos
[?25l Downloading https://files.pythonhosted.org/packages/2a/c0/c3c0ebaa718166af6ff03a4e575d4d1ff4ffbd3ccb658be4285fbeaf3c89/pygeos-0.10-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.whl (2.0MB)
[K |████████████████████████████████| 2.0MB 32.0MB/s
[?25hRequirement already satisfied: joblib in /usr/local/lib/python3.7/dist-packages (from tobler>=0.6.0->pysal) (1.0.1)
Collecting rasterio
[?25l Downloading https://files.pythonhosted.org/packages/0e/78/c69f7457b7dad6163abde2772abd4c8c0c6498d2ab9fd3f3b0eb73b40951/rasterio-1.2.4-cp37-cp37m-manylinux1_x86_64.whl (19.3MB)
[K |████████████████████████████████| 19.3MB 249kB/s
[?25hRequirement already satisfied: statsmodels in /usr/local/lib/python3.7/dist-packages (from tobler>=0.6.0->pysal) (0.10.2)
Collecting rasterstats
Downloading https://files.pythonhosted.org/packages/9f/52/055b2b736e4aa1126c4619a561b44c3bc30fbe48025e6f3275b92928a0a0/rasterstats-0.15.0-py3-none-any.whl
Requirement already satisfied: networkx in /usr/local/lib/python3.7/dist-packages (from mapclassify>=2.4.2->pysal) (2.5.1)
Requirement already satisfied: descartes in /usr/local/lib/python3.7/dist-packages (from splot>=1.1.3->pysal) (1.1.0)
Collecting pulp
[?25l Downloading https://files.pythonhosted.org/packages/14/c4/0eec14a0123209c261de6ff154ef3be5cad3fd557c084f468356662e0585/PuLP-2.4-py3-none-any.whl (40.6MB)
[K |████████████████████████████████| 40.6MB 100kB/s
[?25hRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.7/dist-packages (from python-dateutil<=2.8.0->pysal) (1.15.0)
Requirement already satisfied: setuptools in /usr/local/lib/python3.7/dist-packages (from pytest->pysal) (57.0.0)
Requirement already satisfied: atomicwrites>=1.0 in /usr/local/lib/python3.7/dist-packages (from pytest->pysal) (1.4.0)
Requirement already satisfied: py>=1.5.0 in /usr/local/lib/python3.7/dist-packages (from pytest->pysal) (1.10.0)
Requirement already satisfied: pluggy<0.8,>=0.5 in /usr/local/lib/python3.7/dist-packages (from pytest->pysal) (0.7.1)
Requirement already satisfied: more-itertools>=4.0.0 in /usr/local/lib/python3.7/dist-packages (from pytest->pysal) (8.7.0)
Requirement already satisfied: attrs>=17.4.0 in /usr/local/lib/python3.7/dist-packages (from pytest->pysal) (21.2.0)
Requirement already satisfied: toml in /usr/local/lib/python3.7/dist-packages (from pytest-cov->pysal) (0.10.2)
Requirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->libpysal>=4.4.0->pysal) (2.10)
Requirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->libpysal>=4.4.0->pysal) (2020.12.5)
Requirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->libpysal>=4.4.0->pysal) (3.0.4)
Requirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.7/dist-packages (from jinja2->libpysal>=4.4.0->pysal) (2.0.1)
Requirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.7/dist-packages (from pandas->libpysal>=4.4.0->pysal) (2018.9)
Requirement already satisfied: numba>=0.38 in /usr/local/lib/python3.7/dist-packages (from quantecon>=0.4.7->giddy>=2.3.3->pysal) (0.51.2)
Requirement already satisfied: sympy in /usr/local/lib/python3.7/dist-packages (from quantecon>=0.4.7->giddy>=2.3.3->pysal) (1.7.1)
Requirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->pointpats>=2.2.0->pysal) (1.3.1)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->pointpats>=2.2.0->pysal) (2.4.7)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib->pointpats>=2.2.0->pysal) (0.10.0)
Collecting fiona>=1.8
[?25l Downloading https://files.pythonhosted.org/packages/9c/fc/9807326c37a6bfb2393ae3e1cca147aa74844562c4d5daa782d6e97ad2bc/Fiona-1.8.20-cp37-cp37m-manylinux1_x86_64.whl (15.4MB)
[K |████████████████████████████████| 15.4MB 287kB/s
[?25hCollecting pyproj>=2.2.0
[?25l Downloading https://files.pythonhosted.org/packages/11/1d/1c54c672c2faf08d28fe78e15d664c048f786225bef95ad87b6c435cf69e/pyproj-3.1.0-cp37-cp37m-manylinux2010_x86_64.whl (6.6MB)
[K |████████████████████████████████| 6.6MB 28.4MB/s
[?25hRequirement already satisfied: shapely>=1.6 in /usr/local/lib/python3.7/dist-packages (from geopandas->segregation>=1.5.0->pysal) (1.7.1)
Collecting snuggs>=1.4.1
Downloading https://files.pythonhosted.org/packages/cc/0e/d27d6e806d6c0d1a2cfdc5d1f088e42339a0a54a09c3343f7f81ec8947ea/snuggs-1.4.7-py3-none-any.whl
Collecting affine
Downloading https://files.pythonhosted.org/packages/ac/a6/1a39a1ede71210e3ddaf623982b06ecfc5c5c03741ae659073159184cd3e/affine-2.3.0-py2.py3-none-any.whl
Collecting click-plugins
Downloading https://files.pythonhosted.org/packages/e9/da/824b92d9942f4e472702488857914bdd50f73021efea15b4cad9aca8ecef/click_plugins-1.1.1-py2.py3-none-any.whl
Requirement already satisfied: click>=4.0 in /usr/local/lib/python3.7/dist-packages (from rasterio->tobler>=0.6.0->pysal) (7.1.2)
Collecting cligj>=0.5
Downloading https://files.pythonhosted.org/packages/73/86/43fa9f15c5b9fb6e82620428827cd3c284aa933431405d1bcf5231ae3d3e/cligj-0.7.2-py3-none-any.whl
Requirement already satisfied: patsy>=0.4.0 in /usr/local/lib/python3.7/dist-packages (from statsmodels->tobler>=0.6.0->pysal) (0.5.1)
Collecting simplejson
[?25l Downloading https://files.pythonhosted.org/packages/a8/04/377418ac1e530ce2a196b54c6552c018fdf1fe776718053efb1f216bffcd/simplejson-3.17.2-cp37-cp37m-manylinux2010_x86_64.whl (128kB)
[K |████████████████████████████████| 133kB 51.6MB/s
[?25hRequirement already satisfied: decorator<5,>=4.3 in /usr/local/lib/python3.7/dist-packages (from networkx->mapclassify>=2.4.2->pysal) (4.4.2)
Collecting amply>=0.1.2
Downloading https://files.pythonhosted.org/packages/f3/c5/dfa09dd2595a2ab2ab4e6fa7bebef9565812722e1980d04b0edce5032066/amply-0.1.4-py3-none-any.whl
Requirement already satisfied: llvmlite<0.35,>=0.34.0.dev0 in /usr/local/lib/python3.7/dist-packages (from numba>=0.38->quantecon>=0.4.7->giddy>=2.3.3->pysal) (0.34.0)
Requirement already satisfied: mpmath>=0.19 in /usr/local/lib/python3.7/dist-packages (from sympy->quantecon>=0.4.7->giddy>=2.3.3->pysal) (1.2.1)
Collecting munch
Downloading https://files.pythonhosted.org/packages/cc/ab/85d8da5c9a45e072301beb37ad7f833cd344e04c817d97e0cc75681d248f/munch-2.5.0-py2.py3-none-any.whl
Requirement already satisfied: docutils>=0.3 in /usr/local/lib/python3.7/dist-packages (from amply>=0.1.2->pulp->spopt>=0.1.1->pysal) (0.17.1)
Building wheels for collected packages: pysal, inequality, pointpats, mgwr, spglm, spint, spvcm, splot
Building wheel for pysal (setup.py) ... [?25l[?25hdone
Created wheel for pysal: filename=pysal-2.4.0-cp37-none-any.whl size=18972 sha256=d1f59826eb213aa26ee67e21dda2d040baa6e501f4ce71fab538aa60e9d5efa5
Stored in directory: /root/.cache/pip/wheels/48/c0/2f/ec47118138a00fa2f359e4ad28fe87b393c0c03e2041986ae2
Building wheel for inequality (setup.py) ... [?25l[?25hdone
Created wheel for inequality: filename=inequality-1.0.0-cp37-none-any.whl size=11801 sha256=fd6c08a81c923c2dc73c99dc89b3d8d8a6c3de03a558fec305c34bf0b0c3d21f
Stored in directory: /root/.cache/pip/wheels/0e/cc/44/666696f89f7c7b13cc477bd9c1a9aef180be238550419007f2
Building wheel for pointpats (setup.py) ... [?25l[?25hdone
Created wheel for pointpats: filename=pointpats-2.2.0-cp37-none-any.whl size=60835 sha256=486e35a13c33bb3398e784f359da15bae1cbb6b16d017ccc49ed12c85ef6fc19
Stored in directory: /root/.cache/pip/wheels/f9/28/f2/65c0993e68e25cef954ecd20abff91c6b11f0419f31b60e2ff
Building wheel for mgwr (setup.py) ... [?25l[?25hdone
Created wheel for mgwr: filename=mgwr-2.1.2-cp37-none-any.whl size=46386 sha256=dc34318a7b31136007a7bf256f29df86d83d7b1251e576efc6652d2d592284fe
Stored in directory: /root/.cache/pip/wheels/cf/11/e3/68692f1c637b23a8e2c6e200f6183c3502718b5f7de6dc630d
Building wheel for spglm (setup.py) ... [?25l[?25hdone
Created wheel for spglm: filename=spglm-1.0.8-cp37-none-any.whl size=38806 sha256=044e38098fd853b85197ae738ed36bc117fd40d136ece759c2f6e8b69d607ff4
Stored in directory: /root/.cache/pip/wheels/cd/1e/e3/c68bde79087fdc97cf65ec86e6d1d7ed2171e1baa1b0482363
Building wheel for spint (setup.py) ... [?25l[?25hdone
Created wheel for spint: filename=spint-1.0.7-cp37-none-any.whl size=31384 sha256=fd24fb23f7f64a434442be3b8a0d92393fcf520bf2b639e7629141ed078db745
Stored in directory: /root/.cache/pip/wheels/e1/f0/fe/754baccec76ae29184c37ea6112d881a883e044977bd963d08
Building wheel for spvcm (setup.py) ... [?25l[?25hdone
Created wheel for spvcm: filename=spvcm-0.3.0-cp37-none-any.whl size=5777202 sha256=2d10b78d56842592d4b727dacbd408b186396ab219b032efb2b759780c658e74
Stored in directory: /root/.cache/pip/wheels/8f/2e/c8/35a9abbb5c8508cfc597cdd2217c1b9b2cb82fc2f7fe18c11c
Building wheel for splot (setup.py) ... [?25l[?25hdone
Created wheel for splot: filename=splot-1.1.3-cp37-none-any.whl size=37890 sha256=594b7d9ad27d4a767c102431d25e569cf63ea2f544957daeadb074767228a22e
Stored in directory: /root/.cache/pip/wheels/bd/4f/94/71caf6a544f341a1ef2788a7c96272e7e9dfebb04a1aecc24f
Successfully built pysal inequality pointpats mgwr spglm spint spvcm splot
[31mERROR: requests 2.23.0 has requirement urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1, but you'll have urllib3 1.26.5 which is incompatible.[0m
[31mERROR: datascience 0.10.6 has requirement folium==0.2.1, but you'll have folium 0.8.3 which is incompatible.[0m
[31mERROR: albumentations 0.1.12 has requirement imgaug<0.2.7,>=0.2.5, but you'll have imgaug 0.2.9 which is incompatible.[0m
[31mERROR: pytest-cov 2.12.1 has requirement coverage>=5.2.1, but you'll have coverage 3.7.1 which is incompatible.[0m
[31mERROR: pytest-cov 2.12.1 has requirement pytest>=4.6, but you'll have pytest 3.6.4 which is incompatible.[0m
Installing collected packages: libpysal, access, esda, mapclassify, quantecon, giddy, inequality, opencv-contrib-python, pointpats, click-plugins, munch, cligj, fiona, pyproj, geopandas, segregation, rtree, spaghetti, spreg, spglm, mgwr, spint, spvcm, pygeos, snuggs, affine, rasterio, simplejson, rasterstats, tobler, splot, amply, pulp, spopt, urllib3, python-dateutil, pytest-cov, pysal
Found existing installation: opencv-contrib-python 4.1.2.30
Uninstalling opencv-contrib-python-4.1.2.30:
Successfully uninstalled opencv-contrib-python-4.1.2.30
Found existing installation: urllib3 1.24.3
Uninstalling urllib3-1.24.3:
Successfully uninstalled urllib3-1.24.3
Found existing installation: python-dateutil 2.8.1
Uninstalling python-dateutil-2.8.1:
Successfully uninstalled python-dateutil-2.8.1
Successfully installed access-1.1.3 affine-2.3.0 amply-0.1.4 click-plugins-1.1.1 cligj-0.7.2 esda-2.3.6 fiona-1.8.20 geopandas-0.9.0 giddy-2.3.3 inequality-1.0.0 libpysal-4.4.0 mapclassify-2.4.2 mgwr-2.1.2 munch-2.5.0 opencv-contrib-python-4.5.2.54 pointpats-2.2.0 pulp-2.4 pygeos-0.10 pyproj-3.1.0 pysal-2.4.0 pytest-cov-2.12.1 python-dateutil-2.8.0 quantecon-0.5.0 rasterio-1.2.4 rasterstats-0.15.0 rtree-0.9.7 segregation-1.5.0 simplejson-3.17.2 snuggs-1.4.7 spaghetti-1.6.0 spglm-1.0.8 spint-1.0.7 splot-1.1.3 spopt-0.1.1 spreg-1.2.2 spvcm-0.3.0 tobler-0.7.0 urllib3-1.26.5
Collecting cartoframes
[?25l Downloading https://files.pythonhosted.org/packages/a2/34/e20671c9b56b9d174078fa0556be3b7f848a98174cd5e81e12d8bcc63359/cartoframes-1.2.1-py2.py3-none-any.whl (244kB)
[K |████████████████████████████████| 245kB 33.4MB/s
[?25hRequirement already satisfied: jinja2<3.0,>=2.10.1 in /usr/local/lib/python3.7/dist-packages (from cartoframes) (2.11.3)
Collecting semantic-version<3,>=2.8.0
Downloading https://files.pythonhosted.org/packages/a5/15/00ef3b7888a10363b7c402350eda3acf395ff05bebae312d1296e528516a/semantic_version-2.8.5-py2.py3-none-any.whl
Requirement already satisfied: appdirs<2.0,>=1.4.3 in /usr/local/lib/python3.7/dist-packages (from cartoframes) (1.4.4)
Requirement already satisfied: geopandas<1.0,>=0.6.0 in /usr/local/lib/python3.7/dist-packages (from cartoframes) (0.9.0)
Collecting carto<2.0,>=1.11.2
Downloading https://files.pythonhosted.org/packages/25/e1/d32f14e5827b818aec6509b03fc04867728440035a54878d08c108890a1e/carto-1.11.2-py3-none-any.whl
Collecting unidecode<2.0,>=1.1.0
[?25l Downloading https://files.pythonhosted.org/packages/9e/25/723487ca2a52ebcee88a34d7d1f5a4b80b793f179ee0f62d5371938dfa01/Unidecode-1.2.0-py2.py3-none-any.whl (241kB)
[K |████████████████████████████████| 245kB 46.6MB/s
[?25hRequirement already satisfied: pandas>=0.25.0 in /usr/local/lib/python3.7/dist-packages (from cartoframes) (1.1.5)
Requirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.7/dist-packages (from jinja2<3.0,>=2.10.1->cartoframes) (2.0.1)
Requirement already satisfied: fiona>=1.8 in /usr/local/lib/python3.7/dist-packages (from geopandas<1.0,>=0.6.0->cartoframes) (1.8.20)
Requirement already satisfied: pyproj>=2.2.0 in /usr/local/lib/python3.7/dist-packages (from geopandas<1.0,>=0.6.0->cartoframes) (3.1.0)
Requirement already satisfied: shapely>=1.6 in /usr/local/lib/python3.7/dist-packages (from geopandas<1.0,>=0.6.0->cartoframes) (1.7.1)
Collecting pyrestcli==0.6.11
Downloading https://files.pythonhosted.org/packages/af/24/d3e7638e30f36396caa4ddc1e25d3f1b5ff76d91738166996e74a9cb4fc0/pyrestcli-0.6.11.tar.gz
Requirement already satisfied: requests>=2.7.0 in /usr/local/lib/python3.7/dist-packages (from carto<2.0,>=1.11.2->cartoframes) (2.23.0)
Requirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.7/dist-packages (from pandas>=0.25.0->cartoframes) (2018.9)
Requirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.7/dist-packages (from pandas>=0.25.0->cartoframes) (2.8.0)
Requirement already satisfied: numpy>=1.15.4 in /usr/local/lib/python3.7/dist-packages (from pandas>=0.25.0->cartoframes) (1.19.5)
Requirement already satisfied: click>=4.0 in /usr/local/lib/python3.7/dist-packages (from fiona>=1.8->geopandas<1.0,>=0.6.0->cartoframes) (7.1.2)
Requirement already satisfied: munch in /usr/local/lib/python3.7/dist-packages (from fiona>=1.8->geopandas<1.0,>=0.6.0->cartoframes) (2.5.0)
Requirement already satisfied: click-plugins>=1.0 in /usr/local/lib/python3.7/dist-packages (from fiona>=1.8->geopandas<1.0,>=0.6.0->cartoframes) (1.1.1)
Requirement already satisfied: setuptools in /usr/local/lib/python3.7/dist-packages (from fiona>=1.8->geopandas<1.0,>=0.6.0->cartoframes) (57.0.0)
Requirement already satisfied: cligj>=0.5 in /usr/local/lib/python3.7/dist-packages (from fiona>=1.8->geopandas<1.0,>=0.6.0->cartoframes) (0.7.2)
Requirement already satisfied: certifi in /usr/local/lib/python3.7/dist-packages (from fiona>=1.8->geopandas<1.0,>=0.6.0->cartoframes) (2020.12.5)
Requirement already satisfied: six>=1.7 in /usr/local/lib/python3.7/dist-packages (from fiona>=1.8->geopandas<1.0,>=0.6.0->cartoframes) (1.15.0)
Requirement already satisfied: attrs>=17 in /usr/local/lib/python3.7/dist-packages (from fiona>=1.8->geopandas<1.0,>=0.6.0->cartoframes) (21.2.0)
Requirement already satisfied: future>=0.15.2 in /usr/local/lib/python3.7/dist-packages (from pyrestcli==0.6.11->carto<2.0,>=1.11.2->cartoframes) (0.16.0)
Requirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests>=2.7.0->carto<2.0,>=1.11.2->cartoframes) (2.10)
Requirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests>=2.7.0->carto<2.0,>=1.11.2->cartoframes) (3.0.4)
Collecting urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1
[?25l Downloading https://files.pythonhosted.org/packages/56/aa/4ef5aa67a9a62505db124a5cb5262332d1d4153462eb8fd89c9fa41e5d92/urllib3-1.25.11-py2.py3-none-any.whl (127kB)
[K |████████████████████████████████| 133kB 43.6MB/s
[?25hBuilding wheels for collected packages: pyrestcli
Building wheel for pyrestcli (setup.py) ... [?25l[?25hdone
Created wheel for pyrestcli: filename=pyrestcli-0.6.11-cp37-none-any.whl size=8499 sha256=d2a6a9fdc7900eb4dcd13645cba762251142b431194b90685b25733427c159af
Stored in directory: /root/.cache/pip/wheels/2f/bb/11/396a62e2d1e718f2bfb02b66726240fbc8d98640bfc0cf1688
Successfully built pyrestcli
[31mERROR: pysal 2.4.0 has requirement urllib3>=1.26, but you'll have urllib3 1.25.11 which is incompatible.[0m
[31mERROR: datascience 0.10.6 has requirement folium==0.2.1, but you'll have folium 0.8.3 which is incompatible.[0m
Installing collected packages: semantic-version, pyrestcli, carto, unidecode, cartoframes, urllib3
Found existing installation: urllib3 1.26.5
Uninstalling urllib3-1.26.5:
Successfully uninstalled urllib3-1.26.5
Successfully installed carto-1.11.2 cartoframes-1.2.1 pyrestcli-0.6.11 semantic-version-2.8.5 unidecode-1.2.0 urllib3-1.25.11
Collecting urllib3==1.22
[?25l Downloading https://files.pythonhosted.org/packages/63/cb/6965947c13a94236f6d4b8223e21beb4d576dc72e8130bd7880f600839b8/urllib3-1.22-py2.py3-none-any.whl (132kB)
[K |████████████████████████████████| 133kB 26.8MB/s
[31mERROR: pysal 2.4.0 has requirement urllib3>=1.26, but you'll have urllib3 1.22 which is incompatible.[0m
[31mERROR: datascience 0.10.6 has requirement folium==0.2.1, but you'll have folium 0.8.3 which is incompatible.[0m
[?25hInstalling collected packages: urllib3
Successfully installed urllib3-1.22
Remember to restart the runtime to load correctly the installed libraries!
Now, we download from the web the dataset for this lab.
! wget https://geodacenter.github.io/data-and-lab//data/columbus.zip
! unzip columbus.zip
--2021-06-11 10:11:30-- https://geodacenter.github.io/data-and-lab//data/columbus.zip
Resolving geodacenter.github.io (geodacenter.github.io)... 185.199.108.153, 185.199.109.153, 185.199.110.153, ...
Connecting to geodacenter.github.io (geodacenter.github.io)|185.199.108.153|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 472230 (461K) [application/zip]
Saving to: ‘columbus.zip’
columbus.zip 100%[===================>] 461.16K --.-KB/s in 0.01s
2021-06-11 10:11:30 (43.2 MB/s) - ‘columbus.zip’ saved [472230/472230]
Archive: columbus.zip
creating: columbus/
inflating: columbus/.DS_Store
creating: __MACOSX/
creating: __MACOSX/columbus/
inflating: __MACOSX/columbus/._.DS_Store
inflating: columbus/columbus.csv
inflating: __MACOSX/columbus/._columbus.csv
inflating: columbus/columbus.dbf
inflating: __MACOSX/columbus/._columbus.dbf
creating: columbus/columbus.gdb/
inflating: columbus/columbus.gdb/a00000001.gdbindexes
inflating: columbus/columbus.gdb/a00000001.gdbtable
inflating: columbus/columbus.gdb/a00000001.gdbtablx
inflating: columbus/columbus.gdb/a00000001.TablesByName.atx
inflating: columbus/columbus.gdb/a00000002.gdbtable
inflating: columbus/columbus.gdb/a00000002.gdbtablx
inflating: columbus/columbus.gdb/a00000003.gdbindexes
inflating: columbus/columbus.gdb/a00000003.gdbtable
inflating: columbus/columbus.gdb/a00000003.gdbtablx
inflating: columbus/columbus.gdb/a00000004.CatItemsByPhysicalName.atx
inflating: columbus/columbus.gdb/a00000004.CatItemsByType.atx
inflating: columbus/columbus.gdb/a00000004.FDO_UUID.atx
inflating: columbus/columbus.gdb/a00000004.gdbindexes
inflating: columbus/columbus.gdb/a00000004.gdbtable
inflating: columbus/columbus.gdb/a00000004.gdbtablx
inflating: columbus/columbus.gdb/a00000004.spx
inflating: columbus/columbus.gdb/a00000005.CatItemTypesByName.atx
inflating: columbus/columbus.gdb/a00000005.CatItemTypesByParentTypeID.atx
inflating: columbus/columbus.gdb/a00000005.CatItemTypesByUUID.atx
inflating: columbus/columbus.gdb/a00000005.gdbindexes
inflating: columbus/columbus.gdb/a00000005.gdbtable
inflating: columbus/columbus.gdb/a00000005.gdbtablx
inflating: columbus/columbus.gdb/a00000006.CatRelsByDestinationID.atx
inflating: columbus/columbus.gdb/a00000006.CatRelsByOriginID.atx
inflating: columbus/columbus.gdb/a00000006.CatRelsByType.atx
inflating: columbus/columbus.gdb/a00000006.FDO_UUID.atx
inflating: columbus/columbus.gdb/a00000006.gdbindexes
inflating: columbus/columbus.gdb/a00000006.gdbtable
inflating: columbus/columbus.gdb/a00000006.gdbtablx
inflating: columbus/columbus.gdb/a00000007.CatRelTypesByBackwardLabel.atx
inflating: columbus/columbus.gdb/a00000007.CatRelTypesByDestItemTypeID.atx
inflating: columbus/columbus.gdb/a00000007.CatRelTypesByForwardLabel.atx
inflating: columbus/columbus.gdb/a00000007.CatRelTypesByName.atx
inflating: columbus/columbus.gdb/a00000007.CatRelTypesByOriginItemTypeID.atx
inflating: columbus/columbus.gdb/a00000007.CatRelTypesByUUID.atx
inflating: columbus/columbus.gdb/a00000007.gdbindexes
inflating: columbus/columbus.gdb/a00000007.gdbtable
inflating: columbus/columbus.gdb/a00000007.gdbtablx
inflating: columbus/columbus.gdb/a00000009.gdbindexes
inflating: columbus/columbus.gdb/a00000009.gdbtable
inflating: columbus/columbus.gdb/a00000009.gdbtablx
inflating: columbus/columbus.gdb/a00000009.spx
inflating: columbus/columbus.gdb/gdb
inflating: columbus/columbus.gdb/timestamps
inflating: columbus/columbus.geojson
inflating: columbus/columbus.gpkg
inflating: columbus/columbus.html
inflating: __MACOSX/columbus/._columbus.html
inflating: columbus/columbus.kml
inflating: columbus/columbus.mid
inflating: columbus/columbus.mif
inflating: columbus/columbus.prj
inflating: columbus/columbus.shp
inflating: __MACOSX/columbus/._columbus.shp
inflating: columbus/columbus.shx
inflating: __MACOSX/columbus/._columbus.shx
inflating: columbus/columbus.sqlite
inflating: columbus/columbus.xlsx
inflating: __MACOSX/columbus/._columbus.xlsx
inflating: __MACOSX/._columbus
Let’s start!
Explorative Spatial Data Analysis
import pandas as pd
import numpy as np
import geopandas as gpd
columbus = gpd.read_file("columbus/columbus.shp")
columbus.head()
/usr/local/lib/python3.7/dist-packages/geopandas/_compat.py:110: UserWarning: The Shapely GEOS version (3.8.0-CAPI-1.13.1 ) is incompatible with the GEOS version PyGEOS was compiled with (3.9.1-CAPI-1.14.2). Conversions between both will be slow.
shapely_geos_version, geos_capi_version_string
| AREA | PERIMETER | COLUMBUS_ | COLUMBUS_I | POLYID | NEIG | HOVAL | INC | CRIME | OPEN | PLUMB | DISCBD | X | Y | NSA | NSB | EW | CP | THOUS | NEIGNO | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.309441 | 2.440629 | 2 | 5 | 1 | 5 | 80.467003 | 19.531 | 15.725980 | 2.850747 | 0.217155 | 5.03 | 38.799999 | 44.070000 | 1.0 | 1.0 | 1.0 | 0.0 | 1000.0 | 1005.0 | POLYGON ((8.62413 14.23698, 8.55970 14.74245, ... |
| 1 | 0.259329 | 2.236939 | 3 | 1 | 2 | 1 | 44.567001 | 21.232 | 18.801754 | 5.296720 | 0.320581 | 4.27 | 35.619999 | 42.380001 | 1.0 | 1.0 | 0.0 | 0.0 | 1000.0 | 1001.0 | POLYGON ((8.25279 14.23694, 8.28276 14.22994, ... |
| 2 | 0.192468 | 2.187547 | 4 | 6 | 3 | 6 | 26.350000 | 15.956 | 30.626781 | 4.534649 | 0.374404 | 3.89 | 39.820000 | 41.180000 | 1.0 | 1.0 | 1.0 | 0.0 | 1000.0 | 1006.0 | POLYGON ((8.65331 14.00809, 8.81814 14.00205, ... |
| 3 | 0.083841 | 1.427635 | 5 | 2 | 4 | 2 | 33.200001 | 4.477 | 32.387760 | 0.394427 | 1.186944 | 3.70 | 36.500000 | 40.520000 | 1.0 | 1.0 | 0.0 | 0.0 | 1000.0 | 1002.0 | POLYGON ((8.45950 13.82035, 8.47341 13.83227, ... |
| 4 | 0.488888 | 2.997133 | 6 | 7 | 5 | 7 | 23.225000 | 11.252 | 50.731510 | 0.405664 | 0.624596 | 2.83 | 40.009998 | 38.000000 | 1.0 | 1.0 | 1.0 | 0.0 | 1000.0 | 1007.0 | POLYGON ((8.68527 13.63952, 8.67758 13.72221, ... |
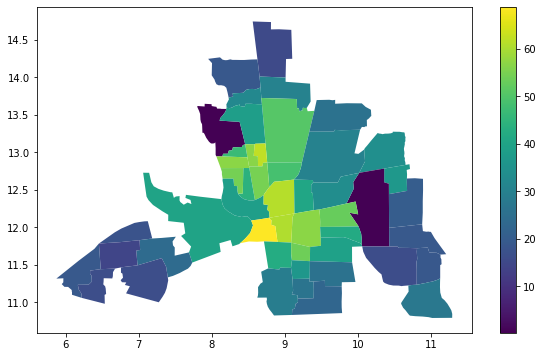
columbus.plot(column="CRIME", figsize=(10, 6),legend=True)
<matplotlib.axes._subplots.AxesSubplot at 0x7f49ab0d0ed0>
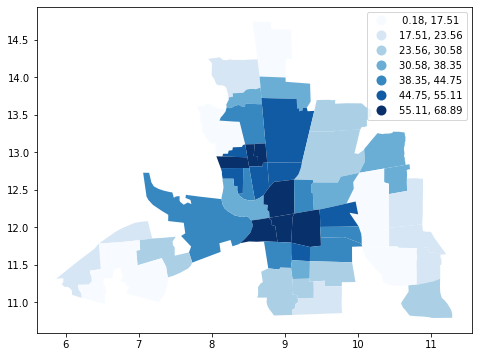
columbus.plot(column="CRIME", figsize=(10, 6), scheme='Quantiles', k=7, cmap='Blues',legend=True);
from cartoframes.viz import Layer, color_bins_style
Layer(
columbus,
color_bins_style('CRIME', palette='CB_BLUES', bins=5, opacity=0.8)
)
NumExpr defaulting to 2 threads.
Let’s create the weight matrix
from libpysal.weights import Queen
w = Queen.from_dataframe(columbus)
w.transform = 'r'
Spatial Lag
Compute the lagged value from the W matrix
$y l a g_{i}=\sum_{j} w_{i, j} y_{j}$
from libpysal.weights import lag_spatial
y = columbus['CRIME']
ylag = lag_spatial(w, y)
ylag
array([24.7142675 , 26.24684033, 29.411751 , 34.64647575, 40.4653275 ,
40.6237135 , 49.728447 , 41.49913083, 36.9477815 , 25.328384 ,
49.857447 , 43.25009117, 38.093982 , 42.82053817, 50.10508383,
52.36547225, 18.09104533, 51.18116975, 46.442163 , 32.4463615 ,
43.72131433, 35.90408433, 18.74605433, 54.90555729, 49.97125037,
48.62930967, 33.20419825, 47.42423011, 51.79066157, 48.5733284 ,
19.12681567, 13.8546865 , 37.27826625, 22.1371835 , 33.098551 ,
18.7549056 , 50.99421883, 43.65270583, 16.171101 , 21.0544898 ,
21.06991733, 19.139792 , 36.16118067, 35.747272 , 32.07386125,
16.7032095 , 17.5732225 , 28.5489635 , 27.21200567])
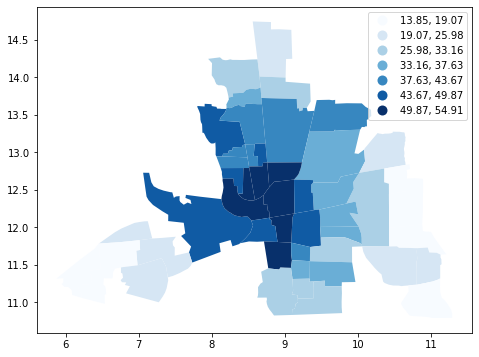
columbus['CRIME_W'] = ylag
columbus.plot(column="CRIME_W", figsize=(10, 6), scheme='Quantiles', k=7, cmap='Blues',legend=True)
<matplotlib.axes._subplots.AxesSubplot at 0x7f4984587e10>
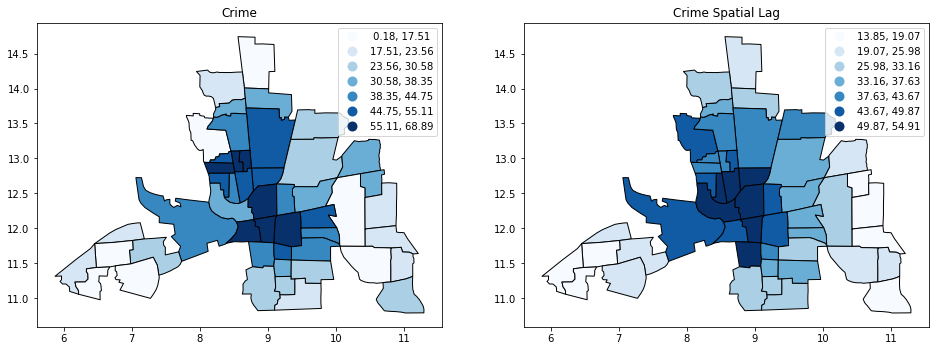
import matplotlib.pyplot as plt
f,ax = plt.subplots(1,2,figsize=(16, 8))
columbus.plot(column='CRIME', ax=ax[0], edgecolor='k',
scheme='Quantiles', k=7, cmap='Blues',legend=True)
ax[0].set_title("Crime")
columbus.plot(column='CRIME_W', ax=ax[1], edgecolor='k',
scheme='Quantiles', k=7, cmap='Blues',legend=True)
ax[1].set_title("Crime Spatial Lag");
Moran’s Index
Now we can compute the Moran’s Index
from esda.moran import Moran
mi = Moran(y, w)
mi.I
0.5001885571828611
import seaborn as sbn
sbn.kdeplot(mi.sim, shade=True)
plt.vlines(mi.I, 0, 1, color='r')
plt.vlines(mi.EI, 0,1)
plt.xlabel("Moran's I");
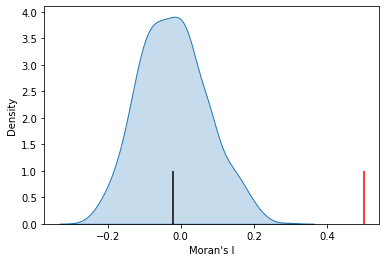
mi.p_norm
1.7988309952698955e-08
mi.p_sim
0.001
https://pysal.org/esda/generated/esda.Moran.html
from splot.esda import moran_scatterplot
moran_scatterplot(mi, aspect_equal=True);
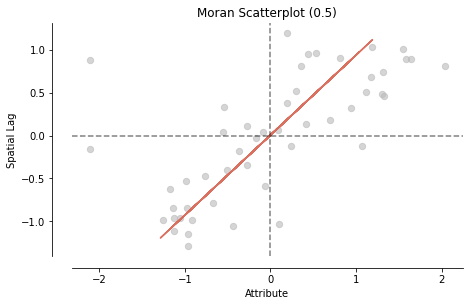
moran_scatterplot(mi, aspect_equal=True, zstandard=False);
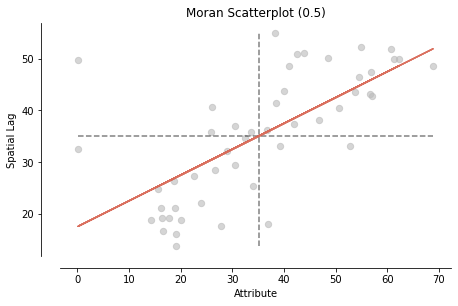
Local Moran
We can also compute the local moran in a similar way
from esda.moran import Moran_Local
mi_local = Moran_Local(y, w)
mi_local.p_sim
array([0.18 , 0.145, 0.232, 0.46 , 0.153, 0.346, 0.055, 0.16 , 0.388,
0.118, 0.014, 0.098, 0.342, 0.11 , 0.007, 0.001, 0.033, 0.029,
0.116, 0.242, 0.185, 0.435, 0.044, 0.001, 0.001, 0.009, 0.442,
0.008, 0.001, 0.015, 0.044, 0.006, 0.391, 0.061, 0.39 , 0.008,
0.006, 0.085, 0.026, 0.018, 0.072, 0.083, 0.426, 0.474, 0.367,
0.048, 0.067, 0.226, 0.206])
moran_scatterplot(mi_local, p=0.05);
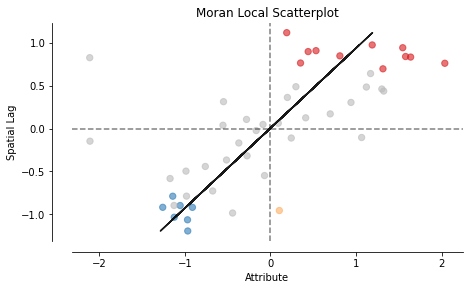
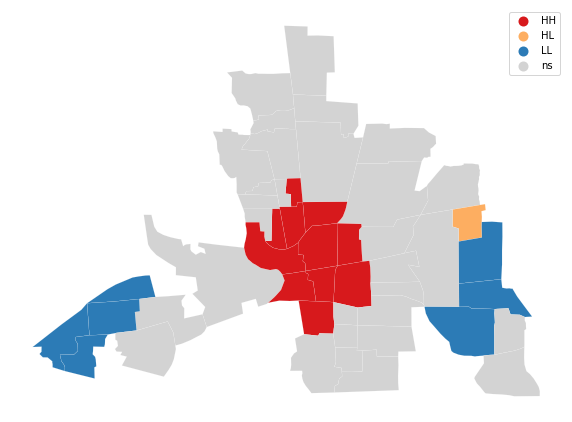
from splot.esda import lisa_cluster
lisa_cluster(mi_local, columbus, p=0.05, figsize = (10,10));
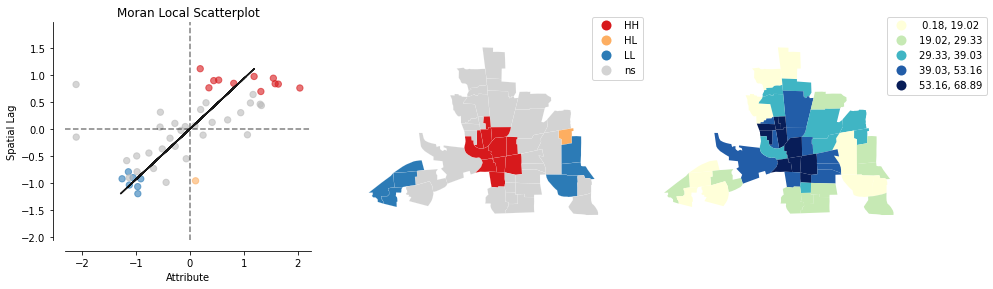
from splot.esda import plot_local_autocorrelation
plot_local_autocorrelation(mi_local, columbus, 'CRIME');
Spatial Econometrics Models
Let’s start with some data manipulation with numpy
columbus['CRIME']
0 15.725980
1 18.801754
2 30.626781
3 32.387760
4 50.731510
5 26.066658
6 0.178269
7 38.425858
8 30.515917
9 34.000835
10 62.275448
11 56.705669
12 46.716129
13 57.066132
14 48.585487
15 54.838711
16 36.868774
17 43.962486
18 54.521965
19 0.223797
20 40.074074
21 33.705048
22 20.048504
23 38.297871
24 61.299175
25 40.969742
26 52.794430
27 56.919785
28 60.750446
29 68.892044
30 17.677214
31 19.145592
32 41.968163
33 23.974028
34 39.175053
35 14.305556
36 42.445076
37 53.710938
38 19.100863
39 16.241299
40 18.905146
41 16.491890
42 36.663612
43 25.962263
44 29.028488
45 16.530533
46 27.822861
47 26.645266
48 22.541491
Name: CRIME, dtype: float64
columbus['CRIME'].values
array([15.72598 , 18.801754, 30.626781, 32.38776 , 50.73151 , 26.066658,
0.178269, 38.425858, 30.515917, 34.000835, 62.275448, 56.705669,
46.716129, 57.066132, 48.585487, 54.838711, 36.868774, 43.962486,
54.521965, 0.223797, 40.074074, 33.705048, 20.048504, 38.297871,
61.299175, 40.969742, 52.79443 , 56.919785, 60.750446, 68.892044,
17.677214, 19.145592, 41.968163, 23.974028, 39.175053, 14.305556,
42.445076, 53.710938, 19.100863, 16.241299, 18.905146, 16.49189 ,
36.663612, 25.962263, 29.028488, 16.530533, 27.822861, 26.645266,
22.541491])
y = columbus['CRIME'].values
y_name = 'CRIME'
x = np.array([columbus.INC]).T
x_name = 'INC'
x
array([[19.531 ],
[21.232 ],
[15.956 ],
[ 4.477 ],
[11.252 ],
[16.028999],
[ 8.438 ],
[11.337 ],
[17.586 ],
[13.598 ],
[ 7.467 ],
[10.048 ],
[ 9.549 ],
[ 9.963 ],
[ 9.873 ],
[ 7.625 ],
[ 9.798 ],
[13.185 ],
[11.618 ],
[31.07 ],
[10.655 ],
[11.709 ],
[21.155001],
[14.236 ],
[ 8.461 ],
[ 8.085 ],
[10.822 ],
[ 7.856 ],
[ 8.681 ],
[13.906 ],
[16.940001],
[18.941999],
[ 9.918 ],
[14.948 ],
[12.814 ],
[18.739 ],
[17.017 ],
[11.107 ],
[18.476999],
[29.833 ],
[22.207001],
[25.872999],
[13.38 ],
[16.961 ],
[14.135 ],
[18.323999],
[18.950001],
[11.813 ],
[18.796 ]])
X_names = ['INC', 'HOVAL']
X = columbus[X_names].values
X
array([[19.531 , 80.467003],
[21.232 , 44.567001],
[15.956 , 26.35 ],
[ 4.477 , 33.200001],
[11.252 , 23.225 ],
[16.028999, 28.75 ],
[ 8.438 , 75. ],
[11.337 , 37.125 ],
[17.586 , 52.599998],
[13.598 , 96.400002],
[ 7.467 , 19.700001],
[10.048 , 19.9 ],
[ 9.549 , 41.700001],
[ 9.963 , 42.900002],
[ 9.873 , 18. ],
[ 7.625 , 18.799999],
[ 9.798 , 41.75 ],
[13.185 , 60. ],
[11.618 , 30.6 ],
[31.07 , 81.266998],
[10.655 , 19.975 ],
[11.709 , 30.450001],
[21.155001, 47.733002],
[14.236 , 53.200001],
[ 8.461 , 17.9 ],
[ 8.085 , 20.299999],
[10.822 , 34.099998],
[ 7.856 , 22.85 ],
[ 8.681 , 32.5 ],
[13.906 , 22.5 ],
[16.940001, 31.799999],
[18.941999, 40.299999],
[ 9.918 , 23.6 ],
[14.948 , 28.450001],
[12.814 , 27. ],
[18.739 , 36.299999],
[17.017 , 43.299999],
[11.107 , 22.700001],
[18.476999, 39.599998],
[29.833 , 61.950001],
[22.207001, 42.099998],
[25.872999, 44.333 ],
[13.38 , 25.700001],
[16.961 , 33.5 ],
[14.135 , 27.733 ],
[18.323999, 76.099998],
[18.950001, 42.5 ],
[11.813 , 26.799999],
[18.796 , 35.799999]])
OLS
from spreg import OLS
ols = OLS(y=y, x=x)
print(ols.summary)
REGRESSION
----------
SUMMARY OF OUTPUT: ORDINARY LEAST SQUARES
-----------------------------------------
Data set : unknown
Weights matrix : None
Dependent Variable : dep_var Number of Observations: 49
Mean dependent var : 35.1288 Number of Variables : 2
S.D. dependent var : 16.7321 Degrees of Freedom : 47
R-squared : 0.4838
Adjusted R-squared : 0.4729
Sum squared residual: 6936.202 F-statistic : 44.0579
Sigma-square : 147.579 Prob(F-statistic) : 2.901e-08
S.E. of regression : 12.148 Log likelihood : -190.869
Sigma-square ML : 141.555 Akaike info criterion : 385.738
S.E of regression ML: 11.8977 Schwarz criterion : 389.521
------------------------------------------------------------------------------------
Variable Coefficient Std.Error t-Statistic Probability
------------------------------------------------------------------------------------
CONSTANT 64.4632283 4.7479549 13.5770516 0.0000000
var_1 -2.0406629 0.3074391 -6.6376157 0.0000000
------------------------------------------------------------------------------------
REGRESSION DIAGNOSTICS
MULTICOLLINEARITY CONDITION NUMBER 5.282
TEST ON NORMALITY OF ERRORS
TEST DF VALUE PROB
Jarque-Bera 2 36.549 0.0000
DIAGNOSTICS FOR HETEROSKEDASTICITY
RANDOM COEFFICIENTS
TEST DF VALUE PROB
Breusch-Pagan test 1 5.072 0.0243
Koenker-Bassett test 1 1.723 0.1893
================================ END OF REPORT =====================================
?ols
ols = OLS(y=y, x=x,
name_y=y_name, name_x=[x_name], name_ds='columbus')
print(ols.summary)
REGRESSION
----------
SUMMARY OF OUTPUT: ORDINARY LEAST SQUARES
-----------------------------------------
Data set : columbus
Weights matrix : None
Dependent Variable : CRIME Number of Observations: 49
Mean dependent var : 35.1288 Number of Variables : 2
S.D. dependent var : 16.7321 Degrees of Freedom : 47
R-squared : 0.4838
Adjusted R-squared : 0.4729
Sum squared residual: 6936.202 F-statistic : 44.0579
Sigma-square : 147.579 Prob(F-statistic) : 2.901e-08
S.E. of regression : 12.148 Log likelihood : -190.869
Sigma-square ML : 141.555 Akaike info criterion : 385.738
S.E of regression ML: 11.8977 Schwarz criterion : 389.521
------------------------------------------------------------------------------------
Variable Coefficient Std.Error t-Statistic Probability
------------------------------------------------------------------------------------
CONSTANT 64.4632283 4.7479549 13.5770516 0.0000000
INC -2.0406629 0.3074391 -6.6376157 0.0000000
------------------------------------------------------------------------------------
REGRESSION DIAGNOSTICS
MULTICOLLINEARITY CONDITION NUMBER 5.282
TEST ON NORMALITY OF ERRORS
TEST DF VALUE PROB
Jarque-Bera 2 36.549 0.0000
DIAGNOSTICS FOR HETEROSKEDASTICITY
RANDOM COEFFICIENTS
TEST DF VALUE PROB
Breusch-Pagan test 1 5.072 0.0243
Koenker-Bassett test 1 1.723 0.1893
================================ END OF REPORT =====================================
Array of betas
ols.betas
array([[64.46322833],
[-2.04066292]])
Array of residuals
ols.u
array([[ -8.88106093],
[ -2.33411931],
[ -1.27562985],
[-22.93942046],
[ 9.2298208 ],
[ -5.6867865 ],
[-47.06584565],
[ -2.90237486],
[ 1.9397867 ],
[ -2.713459 ],
[ 13.04984966],
[ 12.74702165],
[ 1.73919085],
[ 12.9340283 ],
[ 4.26972364],
[ 5.9355374 ],
[ -7.60003908],
[ 6.40539821],
[ 13.76715842],
[ -0.83603454],
[ -2.64589096],
[ -6.86405825],
[ -1.24449831],
[ 2.88551994],
[ 14.1019956 ],
[ -6.99472666],
[ 10.41525574],
[ 8.48800454],
[ 14.00221244],
[ 32.80627417],
[-12.2171825 ],
[ -6.66340142],
[ -2.25577053],
[ -9.98537107],
[ 0.86087927],
[-11.91768995],
[ 12.70780851],
[ 11.91335267],
[ -7.65703868],
[ 12.65716743],
[ -0.24107892],
[ 4.82673124],
[ -0.49554652],
[ -3.88928162],
[ -6.58997002],
[-10.53959011],
[ 2.03019696],
[-13.71161131],
[ -3.56543717]])
Predicted values
ols.predy
array([[24.60704093],
[21.13587331],
[31.90241085],
[55.32718046],
[41.5016892 ],
[31.7534445 ],
[47.24411465],
[41.32823286],
[28.5761303 ],
[36.714294 ],
[49.22559834],
[43.95864735],
[44.97693815],
[44.1321037 ],
[44.31576336],
[48.9031736 ],
[44.46881308],
[37.55708779],
[40.75480658],
[ 1.05983154],
[42.71996496],
[40.56910625],
[21.29300231],
[35.41235106],
[47.1971794 ],
[47.96446866],
[42.37917426],
[48.43178046],
[46.74823356],
[36.08576983],
[29.8943965 ],
[25.80899342],
[44.22393353],
[33.95939907],
[38.31417373],
[26.22324595],
[29.73726749],
[41.79758533],
[26.75790168],
[ 3.58413157],
[19.14622492],
[11.66515876],
[37.15915852],
[29.85154462],
[35.61845802],
[27.07012311],
[25.79266404],
[40.35687731],
[26.10692817]])
$R^2$
ols.r2
0.48384513293999487
Log-likelihood
ols.logll
-190.8688784063158
Jarque Bera
ols.jarque_bera
{'df': 2, 'jb': 36.5488718687176, 'pvalue': 1.1574795154584871e-08}
https://spreg.readthedocs.io/en/latest/generated/spreg.OLS.html
Moran’s I on residual
Using the W matrix in combination with the OLS is possible to compute the Moran’s I on the residuals
from spreg import MoranRes
m = MoranRes(ols, w, z=True)
Value of the Moran’s I statistic:
print(round(m.I,4))
0.1724
Value of the Moran’s I expectation:
print(round(m.eI,4))
-0.0301
Value of the Moran’s I variance:
print(round(m.vI,4))
0.0082
Value of the Moran’s I standardized value. This is distributed as a standard Normal(0, 1)
print(round(m.zI,4))
2.237
P-value of the standardized Moran’s I value (z):
print(round(m.p_norm,4))
0.0253
Otherwise, the preferred approach is to perform all the test usign OLS function:
ols = OLS(y=y, x=x, w=w,
name_y=y_name, name_x = [x_name], name_ds='columbus',
white_test=True, spat_diag=True, moran=True)
print(ols.summary)
REGRESSION
----------
SUMMARY OF OUTPUT: ORDINARY LEAST SQUARES
-----------------------------------------
Data set : columbus
Weights matrix : unknown
Dependent Variable : CRIME Number of Observations: 49
Mean dependent var : 35.1288 Number of Variables : 2
S.D. dependent var : 16.7321 Degrees of Freedom : 47
R-squared : 0.4838
Adjusted R-squared : 0.4729
Sum squared residual: 6936.202 F-statistic : 44.0579
Sigma-square : 147.579 Prob(F-statistic) : 2.901e-08
S.E. of regression : 12.148 Log likelihood : -190.869
Sigma-square ML : 141.555 Akaike info criterion : 385.738
S.E of regression ML: 11.8977 Schwarz criterion : 389.521
------------------------------------------------------------------------------------
Variable Coefficient Std.Error t-Statistic Probability
------------------------------------------------------------------------------------
CONSTANT 64.4632283 4.7479549 13.5770516 0.0000000
INC -2.0406629 0.3074391 -6.6376157 0.0000000
------------------------------------------------------------------------------------
REGRESSION DIAGNOSTICS
MULTICOLLINEARITY CONDITION NUMBER 5.282
TEST ON NORMALITY OF ERRORS
TEST DF VALUE PROB
Jarque-Bera 2 36.549 0.0000
DIAGNOSTICS FOR HETEROSKEDASTICITY
RANDOM COEFFICIENTS
TEST DF VALUE PROB
Breusch-Pagan test 1 5.072 0.0243
Koenker-Bassett test 1 1.723 0.1893
SPECIFICATION ROBUST TEST
TEST DF VALUE PROB
White 2 3.831 0.1473
DIAGNOSTICS FOR SPATIAL DEPENDENCE
TEST MI/DF VALUE PROB
Moran's I (error) 0.1724 2.237 0.0253
Lagrange Multiplier (lag) 1 7.777 0.0053
Robust LM (lag) 1 4.890 0.0270
Lagrange Multiplier (error) 1 3.137 0.0765
Robust LM (error) 1 0.250 0.6170
Lagrange Multiplier (SARMA) 2 8.027 0.0181
================================ END OF REPORT =====================================
ols = OLS(y=y, x=X, w=w,
name_y=y_name, name_x = X_names, name_w="Q row std", name_ds='columbus',
white_test=True, spat_diag=True, moran=True)
print(ols.summary)
REGRESSION
----------
SUMMARY OF OUTPUT: ORDINARY LEAST SQUARES
-----------------------------------------
Data set : columbus
Weights matrix : Q row std
Dependent Variable : CRIME Number of Observations: 49
Mean dependent var : 35.1288 Number of Variables : 3
S.D. dependent var : 16.7321 Degrees of Freedom : 46
R-squared : 0.5524
Adjusted R-squared : 0.5329
Sum squared residual: 6014.893 F-statistic : 28.3856
Sigma-square : 130.759 Prob(F-statistic) : 9.341e-09
S.E. of regression : 11.435 Log likelihood : -187.377
Sigma-square ML : 122.753 Akaike info criterion : 380.754
S.E of regression ML: 11.0794 Schwarz criterion : 386.430
------------------------------------------------------------------------------------
Variable Coefficient Std.Error t-Statistic Probability
------------------------------------------------------------------------------------
CONSTANT 68.6189611 4.7354861 14.4903731 0.0000000
INC -1.5973108 0.3341308 -4.7804962 0.0000183
HOVAL -0.2739315 0.1031987 -2.6544086 0.0108745
------------------------------------------------------------------------------------
REGRESSION DIAGNOSTICS
MULTICOLLINEARITY CONDITION NUMBER 6.542
TEST ON NORMALITY OF ERRORS
TEST DF VALUE PROB
Jarque-Bera 2 1.836 0.3994
DIAGNOSTICS FOR HETEROSKEDASTICITY
RANDOM COEFFICIENTS
TEST DF VALUE PROB
Breusch-Pagan test 2 7.900 0.0193
Koenker-Bassett test 2 5.694 0.0580
SPECIFICATION ROBUST TEST
TEST DF VALUE PROB
White 5 19.946 0.0013
DIAGNOSTICS FOR SPATIAL DEPENDENCE
TEST MI/DF VALUE PROB
Moran's I (error) 0.2221 2.839 0.0045
Lagrange Multiplier (lag) 1 8.898 0.0029
Robust LM (lag) 1 3.736 0.0533
Lagrange Multiplier (error) 1 5.206 0.0225
Robust LM (error) 1 0.044 0.8340
Lagrange Multiplier (SARMA) 2 8.942 0.0114
================================ END OF REPORT =====================================
How to select the spatial specification?
Spatial specification search (Anselin, 2015)
“However, this decision rule needs to be applied with caution, since there may not be sufficient evidence to clearly select one alternative over another”
Spatial Lag
Reference: Anselin, 1998
from spreg import ML_Lag
lag = ML_Lag(y=y, x=X, w=w,
name_y=y_name, name_x = X_names, name_w="Q row std", name_ds='columbus')
print(lag.summary)
REGRESSION
----------
SUMMARY OF OUTPUT: MAXIMUM LIKELIHOOD SPATIAL LAG (METHOD = FULL)
-----------------------------------------------------------------
Data set : columbus
Weights matrix : Q row std
Dependent Variable : CRIME Number of Observations: 49
Mean dependent var : 35.1288 Number of Variables : 4
S.D. dependent var : 16.7321 Degrees of Freedom : 45
Pseudo R-squared : 0.6473
Spatial Pseudo R-squared: 0.5806
Sigma-square ML : 96.857 Log likelihood : -182.674
S.E of regression : 9.842 Akaike info criterion : 373.348
Schwarz criterion : 380.915
------------------------------------------------------------------------------------
Variable Coefficient Std.Error z-Statistic Probability
------------------------------------------------------------------------------------
CONSTANT 45.6032490 7.2574039 6.2836862 0.0000000
INC -1.0487282 0.3074059 -3.4115419 0.0006460
HOVAL -0.2663348 0.0890963 -2.9892918 0.0027962
W_CRIME 0.4233254 0.1195104 3.5421625 0.0003969
------------------------------------------------------------------------------------
================================ END OF REPORT =====================================
/usr/local/lib/python3.7/dist-packages/scipy/optimize/_minimize.py:770: RuntimeWarning: Method 'bounded' does not support relative tolerance in x; defaulting to absolute tolerance.
"defaulting to absolute tolerance.", RuntimeWarning)
/usr/local/lib/python3.7/dist-packages/numpy/core/fromnumeric.py:87: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray
return ufunc.reduce(obj, axis, dtype, out, **passkwargs)
from spreg import GM_Lag
lag = GM_Lag(y=y, x=X, w=w,
name_y=y_name, name_x = X_names, name_w="Q row std", name_ds='columbus')
print(lag.summary)
REGRESSION
----------
SUMMARY OF OUTPUT: SPATIAL TWO STAGE LEAST SQUARES
--------------------------------------------------
Data set : columbus
Weights matrix : Q row std
Dependent Variable : CRIME Number of Observations: 49
Mean dependent var : 35.1288 Number of Variables : 4
S.D. dependent var : 16.7321 Degrees of Freedom : 45
Pseudo R-squared : 0.6494
Spatial Pseudo R-squared: 0.5786
------------------------------------------------------------------------------------
Variable Coefficient Std.Error z-Statistic Probability
------------------------------------------------------------------------------------
CONSTANT 43.9631909 10.7680847 4.0827308 0.0000445
INC -1.0096372 0.3723948 -2.7112007 0.0067040
HOVAL -0.2657935 0.0886026 -2.9998391 0.0027012
W_CRIME 0.4534908 0.1834172 2.4724546 0.0134189
------------------------------------------------------------------------------------
Instrumented: W_CRIME
Instruments: W_HOVAL, W_INC
================================ END OF REPORT =====================================
Spatial Error
Reference: Anselin, 1998
from spreg import ML_Error
error = ML_Error(y=y, x=X, w=w,
name_y=y_name, name_x = X_names, name_w="Q row std", name_ds='columbus')
print(error.summary)
REGRESSION
----------
SUMMARY OF OUTPUT: MAXIMUM LIKELIHOOD SPATIAL ERROR (METHOD = FULL)
-------------------------------------------------------------------
Data set : columbus
Weights matrix : Q row std
Dependent Variable : CRIME Number of Observations: 49
Mean dependent var : 35.1288 Number of Variables : 3
S.D. dependent var : 16.7321 Degrees of Freedom : 46
Pseudo R-squared : 0.5362
Sigma-square ML : 97.674 Log likelihood : -183.749
S.E of regression : 9.883 Akaike info criterion : 373.499
Schwarz criterion : 379.174
------------------------------------------------------------------------------------
Variable Coefficient Std.Error z-Statistic Probability
------------------------------------------------------------------------------------
CONSTANT 60.2794691 5.3655939 11.2344448 0.0000000
INC -0.9573053 0.3342308 -2.8642047 0.0041806
HOVAL -0.3045593 0.0920473 -3.3087251 0.0009372
lambda 0.5467531 0.1380508 3.9605217 0.0000748
------------------------------------------------------------------------------------
================================ END OF REPORT =====================================
/usr/local/lib/python3.7/dist-packages/scipy/optimize/_minimize.py:770: RuntimeWarning: Method 'bounded' does not support relative tolerance in x; defaulting to absolute tolerance.
"defaulting to absolute tolerance.", RuntimeWarning)
Reference: Kelejian and Prucha (1998, 1999)
from spreg import GM_Error
error = GM_Error(y=y, x=X, w=w,
name_y=y_name, name_x = X_names, name_w="Q row std", name_ds='columbus')
print(error.summary)
REGRESSION
----------
SUMMARY OF OUTPUT: SPATIALLY WEIGHTED LEAST SQUARES
---------------------------------------------------
Data set : columbus
Weights matrix : Q row std
Dependent Variable : CRIME Number of Observations: 49
Mean dependent var : 35.1288 Number of Variables : 3
S.D. dependent var : 16.7321 Degrees of Freedom : 46
Pseudo R-squared : 0.5453
------------------------------------------------------------------------------------
Variable Coefficient Std.Error z-Statistic Probability
------------------------------------------------------------------------------------
CONSTANT 62.9187786 5.0108898 12.5564083 0.0000000
INC -1.1500724 0.3347166 -3.4359582 0.0005905
HOVAL -0.2982308 0.0948124 -3.1454814 0.0016581
lambda 0.3834563
------------------------------------------------------------------------------------
================================ END OF REPORT =====================================
Lag + Error
Reference: Kelejian and Prucha (1998, 1999)
from spreg import GM_Combo
combo = GM_Combo(y=y, x=X, w=w,
name_y=y_name, name_x = X_names, name_w="Q row std", name_ds='columbus')
print(combo.summary)
REGRESSION
----------
SUMMARY OF OUTPUT: SPATIALLY WEIGHTED TWO STAGE LEAST SQUARES
-------------------------------------------------------------
Data set : columbus
Weights matrix : Q row std
Dependent Variable : CRIME Number of Observations: 49
Mean dependent var : 35.1288 Number of Variables : 4
S.D. dependent var : 16.7321 Degrees of Freedom : 45
Pseudo R-squared : 0.6494
Spatial Pseudo R-squared: 0.5787
------------------------------------------------------------------------------------
Variable Coefficient Std.Error z-Statistic Probability
------------------------------------------------------------------------------------
CONSTANT 43.9717784 10.7831309 4.0778303 0.0000455
INC -1.0121280 0.3729276 -2.7140070 0.0066475
HOVAL -0.2651583 0.0885408 -2.9947599 0.0027466
W_CRIME 0.4535726 0.1832568 2.4750651 0.0133212
lambda -0.0069612
------------------------------------------------------------------------------------
Instrumented: W_CRIME
Instruments: W_HOVAL, W_INC
================================ END OF REPORT =====================================
combo